Introduction
Pf3k is an international collaboration using the latest sequencing technologies to provide a high-resolution view of natural variation in the malaria parasite Plasmodium falciparum.
The Pf3k project is led by researchers at the Broad Institute, the University of Oxford and the Wellcome Trust Sanger Institute.
Our primary goal is to undertake a comprehensive analysis of genome variation in 3,000 parasite samples representing the major malaria endemic regions of the world. In doing so, we'll:
- Provide an open set of P. falciparum genome sequence data that captures common variation across multiple populations in different parts of the world
- Use a combination of short- and long-read sequencing technologies in controlled settings to establish standards for accuracy and completeness in the inference of P. falciparum genome sequence variation and to characterise the quality of information obtained from standard approaches
- Combine information from read-mapping, full de novo assembly, variant assembly and iterative reassembly of specific genes to obtain the most comprehensive resource on P. falciparum variation to date
- Develop new high-quality reference genomes that will increase the resolution and accuracy of variation analysis across the whole sample set
- Analyse the data to learn about parasite population structure, epidemiology and history, mutational and recombinational processes generating diversity, evolutionary processes including drug resistance and immune evasion, and how such phenomena differ between populations and regions
The primary output of the project will be an open access data resource with companion publications on genomic diversity and population genetics that together provide a detailed description of P. falciparum genome variation across the major malaria endemic regions.
Other outputs will include papers on methodology and standardisation of protocols for P. falciparum sequence analysis and genotyping calling. All of the underlying data will be made publicly available for use by the scientific community, initially under Fort Lauderdale conditions.
Scientific working groups will drive forward specific areas of analysis including statistics and population genetics (led by Gil McVean and Roberto Amato), technology benchmarking (led by Dan Neafsey and Jim Stalker) and reference genomes (led by Matt Berriman and Thomas Otto). The MalariaGEN Resource Centre will provide support for partner studies, data production pipelines, communications and project management. The Project is overseen by the Pf3k Management Committee that is comprised of working group leaders, with support from members of the MalariaGEN Resource Centre.
Pilot phase
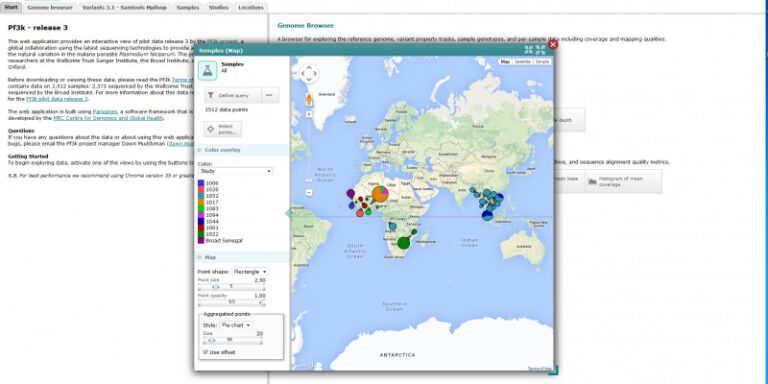
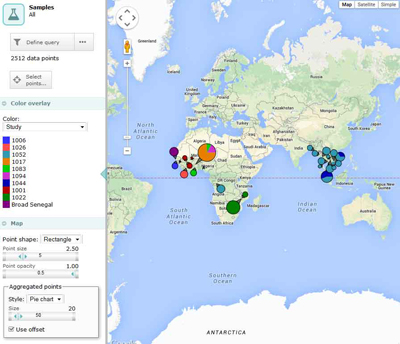
The Pf3k project will have several discrete phases, beginning with a pilot phase which commenced in June 2014. During the pilot phase, the Project is analysing Illumina short-read sequence data on 2,512 samples from multiple locations in Africa and Asia, together with laboratory samples for benchmarking and methods development. The MalariaGEN P. falciparum Community Project and the Broad Institute, together with their partners, have contributed the samples for the pilot phase. The Project will generate genotype calls by a range of different methods, and will perform methodological comparisons and performance metrics.
Planned analyses
During the pilot phase, the Project is undertaking a series of planned analyses that will form the basis of a manuscript, 'A global reference for genomic variation in Plasmodium falciparum', using Pilot Phase data (2,512 samples).
- Sequence data and quality including SNPs, short tandem repeats, haplotypes and patterns of linkage disequilibrium
- Population genetic phenomena such as population comparisons, mutation and recombination rates (haplotype structure and LD)
- Signals of selection and demographic analyses
- Merozoite surface proteins
- var genes and genes implicated in drug resistance
Removing Pf3k Pilot Phase Terms of Use
The Pf3K Pilot Phase Terms of Use were applied to Pilot Phase data releases when they were publically released. In September 2016 these restrictions have been lifted from Pf3k pilot data release packages 1-5 and the data are available open access.
Sampling locations
Bangladesh, Cambodia, Congo, The Gambia, Ghana, Guinea, Laos, Malawi, Mali, Myanmar, Nigeria, Senegal, Thailand, Vietnam.
Data
The Project will publicly release data on a regular basis and prior to publication. Raw sequence reads will be deposited in either the European Nucleotide Archive (ENA) or the NCBI. Alignments and variant calls will be released on individual samples, and data formats and software developed by the Project will be made publicly available. Associated sample information will be made available in the public domain through the MalariaGEN website and other public databases as appropriate. Public release of the data will be associated with contact information for the lead investigators that have contributed the samples.
At the time of their release, these data were subject to the Pf3k Pilot Phase Terms of Use. In September 2016, these restrictions were lifted and this dataset is now available open access.
Current
Sample set: 2,512 field isolates; 5 lab clonal samples; 96 crosses samples; 27 mixed lab strains
Sample information, accession numbers, analysis BAMs, and a set of de novo genotype calls, both indels and SNPs, built using a pipeline based on GATK best practices
This data is
Open access
Sample set: 2,517 samples from 14 countries and 5 lab strains (7G8, GB4, KH02, KE01, GN01)
Sample information, analysis BAMs, and de novo genotype calls built using a pipeline based on GATK best practices
This data is
Open access
Archived
Sample set: 2,512 samples from 14 countries
Sample information, accession numbers, and genotypes
This data is
Open access
Archived
Sample set: 1,931 samples from 12 countries
Sample information, accession numbers, and genotypes
This data is
Open access
Archived
Sample set: 1,794 samples from 11 countries
Sample information and accession numbers
This data is
Open access
Archived
Partner studies
We work with investigators who are pursuing independent partner studies in a number of malaria-endemic countries. Click a link below to learn more about their work.
Partner study description Abdoulaye Djimde and his colleagues worked with MalariaGEN to collect clinical parasite samples from three sites in Mali: Bamako, the capital, and Kolle and Faladje, rural villages approximately 60km and…
Partner study description Alfred Amambua-Ngwa, David Conway, and colleagues surveyed clinical Plasmodium falciparum isolates from The Gambia to assess several measures of genetic variation including allele frequency spectra and signatures of balancing selection,…
Partner study description In collaboration with colleagues at the Navrongo Health Research Center, Dr Lucas Amenga-Etego conducted his thesis research under the guidance of Prof Dominic Kwiatkowski on genetic diversity in natural populations…
Partner study description As part of his PhD research Harold Ocholla worked with colleagues at the Liverpool School of Tropical Medicine and in Malawi to collect uncultured paediatric Plasmodium falciparum isolates from malaria…
Partner study description This study was designed to identify malaria parasite genes under selection in the highly endemic forested area of southern Guinea, where very few studies on malaria have been conducted previously.…
Partner study description In field-based studies, Rick Fairhurst and colleagues investigated patient responses to artemisinin combination therapies (ACTs), in three Cambodian provinces, where artemisinin resistance is entrenched (Pursat), emerging (Preah Vihear), or uncommon…
Partner study description TRAC is investigating the scope and spread of parasite resistance to artemisinin-based therapies at sites across Asia and Africa. The first TRAC study has been completed. This multi-centre, open-label randomised…
Partner study description The intensity of malaria transmission varies considerably among sites in Ghana due to differences in average temperatures, rainfall patterns, and urbanization. This study has selected two locations that have among…
Partner study description In collaboration with colleagues at the Navrongo Health Research Center, Lucas Amenga-Etego is investigating the genetic diversity and population structure of Plasmodium falciparum parasites collected in the Kassena-Nankana districts of…
P. falciparum samples were collected between 2001 and 2011, and were sequenced via a variety of methods: following culture adaption, directly from patient samples without WBC depletion, and from patient samples following hybrid…
Publications
- Genomic analysis of Indian isolates of Plasmodium falciparum: Implications for drug resistance and virulence factors
Choubey et alInt J Parasitol Drugs Drug Resist, 2023; 22 52–60- Population dynamics and drug resistance mutations in Plasmodium falciparum on the Bijagós Archipelago, Guinea-Bissau
Moss et alScientific Reports, 2023; 13(1) 6311- Characterization of a novel Plasmodium falciparum merozoite surface antigen and potential vaccine target
Niaré et alFrontiers Immunology, 2023; 14 1156806- Potent acyl-CoA synthetase 10 inhibitors kill Plasmodium falciparum by disrupting triglyceride formation
Bopp et alNat Commun, 2023; 14(1455)- Genetic diversity and natural selection of rif gene (PF3D7_1254800) in the Plasmodium falciparum global populations
Xu et alMol Biochem Parasitol, 2023; 254 111558- Short tandem repeat polymorphism in the promoter region of cyclophilin 19B drives its transcriptional upregulation and contributes to drug resistance in the malaria parasite Plasmodium falciparum
Kucharski M et alPLOS Pathogens, 2023; 19(1) e1011118- Pf7: An open dataset of Plasmodium falciparum genome variation in 20,000 worldwide samples
MalariaGEN et. alWellcome Open Research, 2023; 8 22- Unravelling var complexity: Relationship between DBLα types and var genes in Plasmodium falciparum
Tan et alFront Parasitol, 2023; 1 1006341- Deconvolution of multiple infections in Plasmodium falciparum from high throughput sequencing data
Zhu, Almagro-Garcia, McVean.Bioinformatics, 2017;
2,512 samples
from
14 countries
Project contact
People
Investigators involved in the Pf3k pilot phase include:
- Abdoulaye Djimdé
- Prof Alfred Amambua-Ngwa
- Alistair Miles
- Prof Alister Craig
- Prof Arjen Dondorp
- Prof Chris Newbold
- Daniel Neafsey
- Prof David Conway
- Dr Marcelo Ferreira
- Dominic Kwiatkowski
- Prof Nadira Karunaweera
- Prof Gilean McVean
- Dr Sarah Auburn
- Prof Gordon Awandare
- Hema Sharma
- Dr John O'Brien
- Dr Joe Zhu
- Dr Lucas Amenga-Etego
- Dr Matt Berriman
- Prof Nicholas J White
- Prof Nicholas Day
- Olivo Miotto
- Richard Pearson
- Dr Rick Fairhurst
- Dr Roberto Amato
- Sorina Maciuca
- Dr Deus Ishengoma
- Dr Thomas D Otto
- Dr Zamin Iqbal
Updates
26 Sep 2016
Pilot release 5 open access
The terms of use applied at the time of release to the Pf3k pilot release 5, and all previous releases, have now been removed and these data are available open access.
9 Feb 2016
Pf3k pilot release 5 is online
We've made our fifth public data release including de novo variant discovery and genotyping across 2,640 P. falciparum samples. The updated sample set now also includes a number of crosses samples and mixed lab strains. The genotyping, both indel and SNP variants, was performed using a pipeline based on GATK best practices. Download the data via FTP.
16 Oct 2015
Pf3k pilot data release 4 is online
We've made our fourth public data release, this time including de novo variant discovery and genotyping across 2,512 samples collected in 14 countries, as well as five lab strains included for method development validation. The genotyping, including both indel and SNP variants, was performed using a pipeline based on GATK best practices. Download the data via FTP.