Introduction
Ag1000G is an international collaboration using whole genome deep sequencing to provide a high-resolution view of genetic variation in natural populations of Anopheles gambiae, the principal vector of Plasmodium falciparum malaria in Africa.
Objectives & Coordination
We have three core objectives:
- Discovering natural genetic variation – We’re using high-throughput sequencing of a large number of wild-caught mosquitoes sampled from across Africa to build a comprehensive catalogue of genetic variation in natural vector populations. Our primary focus is on A. gambiae sensu strictu and A. coluzzii, but we will be expanding to include A. arabiensis in the future.
- Describing the structure and history of vector populations – We’re analysing genetic variation data to characterise key features of natural vector populations, such as patterns of diversity, linkage disequilibrium and recombination, population structure and gene flow, signals of recent selection, and demographic history.
- Connecting genetic variation and population biology with ecology and malaria epidemiology – We aim to study associations between genotype and broad phenotypes such as ecological specialisation and differences in local malaria epidemiology.
Our work is coordinated by two working groups:
- Partner Working Group (led by Martin Donnelly) is comprised of the investigators contributing samples or other significant resources and/or strategic guidance
- Data Analysis Group (led by Dominic Kwiatkowski) is undertaking core project analyses
Locations
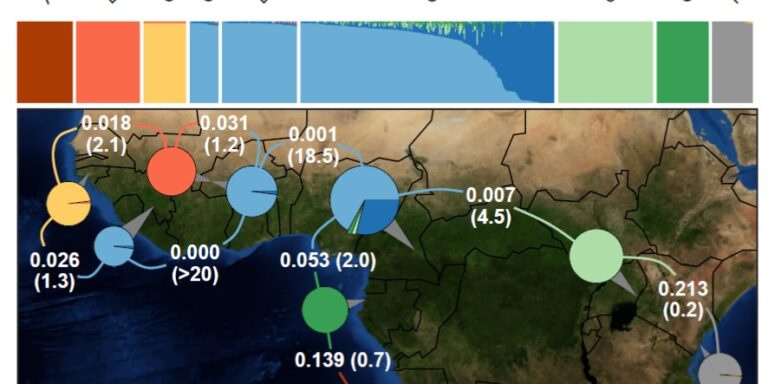
To date, we’ve collected and sequenced mosquitoes from 13 countries:
Sampling locations
Angola, Bioko Island, Equatorial Guinea, Burkina Faso, Cameroon, Central African Republic, Congo, Equatorial Guinea, Gabon, The Gambia, Ivory Coast, Kenya, Malawi, Mali, Mayotte Island, Mozambique, Tanzania, Uganda.
Data
Ag1000G samples are sequenced by the Wellcome Trust Sanger Institute Parasites and Microbes Programme using Illumina high-throughput technology. The sequence data are then used to discover genetic variation between samples and to make genotype calls. Raw sequence reads are deposited into the European Nucleotide Archive (ENA). The variant call data generated by the project is publicly released on a regular basis.
Any use of Ag1000G data is subject to the Terms of Use.
Current
Phased haplotypes for 2,784 wild-caught mosquitoes collected from 19 countries in sub-Saharan Africa.
This data is subject to
Ag1000G Terms of Use
Sample set: 2,784 wild-caught specimens from 19 countries and 297 specimens comprising parents and progeny of 15 crosses.
This data is subject to
Ag1000G Terms of Use
Sample set: 2,784 wild-caught specimens from 19 countries and 297 specimens comprising parents and progeny of 15 crosses.
This data is subject to
Ag1000G Terms of Use
Sample set: 1,142 wild-caught specimens from 13 countries and 234 specimens comprising parents and progeny of 11 crosses.
This data is subject to
Open access
Sample set: 765 wild-caught specimens from 8 countries and 80 specimens comprising parents and progeny of four crosses.
This data is subject to
Open access
Archived
Sample set: 765 wild-caught specimens from 8 countries and 80 specimens comprising parents and progeny of four crosses
This data is subject to
Open access
Archived
Sample set: 765 wild-caught specimens from 8 countries
This data is subject to
Open access
Archived
Sample set: 103 wild-caught specimens from Uganda
This data is subject to
Open access
Archived
For partners
Resources for partners working on mosquitoes
Partner studies
The Ag1000G project is coordinated by a consortium of partners from a range of different research institutions and countries. This includes consortium members who are carrying out independent research studies in malaria endemic regions, and who have contributed mosquito specimens or mosquito DNA samples collected in the course of their own research. The pages below describe the studies that have contributed samples to the Ag1000G project, which includes wild-caught samples from 19 African countries. They also provide information about the collection locations and methods, the people involved in the studies, and references to any published articles providing further information about the studies.
We use species nomenclature following (1). Unless otherwise stated, the DNA extraction method used for the collections described below was Qiagen DNeasy Blood and Tissue Kit (Qiagen Science, MD, USA).
1. Maureen Coetzee, Richard H. Hunt, Richard Wilkerson, Alessandra Della Torre, Mamadou B. Coulibaly, and Nora J. Besansky. Anopheles coluzzii and anopheles amharicus, new members of the anopheles gambiae complex. Zootaxa, 3619(3):246–274, February 2013. doi:10.11646/zootaxa.3619.3.2.
Partner study description Adult mosquitoes were obtained by rearing larvae collected from breeding sites along the main roads connecting the municipalities of Kilamba-Kiaxi and Viana, Luanda province (-8.821, 13.291), in April/May 2009. These…
Partner study description The Target Malaria project contributed samples from collections made in three villages separated by at most 30km: Bana (11.233, -4.472), Souroukoudinga (11.235, -4.535) and Pala (11.150, -4.235). These collections were…
Partner study description Samples were contributed from collections of indoor resting adults made by spray catch from Monomtenga in central Burkina Faso (12.06, -1.17). These specimens were sorted morphologically to An. gambiae s.l..…
Partner study description Pyrethrum spray collections were conducted in three villages in Cameroon during September and October 2009. These villages comprise a transect from forest (village of Mayos: 4.341, 13.558) to forest/savanna transition…
Partner study description These samples were collected as part of a study which took place in Cameroon in Central Africa. The country is commonly referred to as “miniature Africa”, owing to the diversity…
Partner study description Samples were contributed from pyrethrum spray collections, larval sampling and human landing catches conducted in twenty locations during October 2013. These villages are scattered throughout the country and reflect a…
Partner study description Collections were carried out in Bangui (4.367, 18.583), during December 1993, by indoor resting aspiration or pyrethrum spray catch. Contributors Alessandra della Torre (alessandra.dellatorre@uniroma1.it) Istituto Pasteur Italia–Fondazione Cenci Bolognetti, Dipartimento…
Partner study description 15 crosses were contributed to Ag1000G phase 3, 11 of which were previously released in Ag1000G phase 2. Crosses were generated using parents from eight different colonies: G3 (MRA-112); Kisumu…
Partner study description Samples were collected in Tiassale (5.898, -4.823), located in the evergreen forest zone of southern Côte d’Ivoire. The primary agricultural activity is rice cultivation in irrigated fields. High malaria transmission…
Partner study description Samples were collected from Gbadolite (4.283, 21.017), a town located in the far north of the Democratic Republic of Congo (DRC) near the border with the Central African Republic, surrounded…
Partner study description Collections were performed during the rainy season in September 2002 by overnight CDC light traps in Sacriba of Bioko island (3.7, 8.7). Specimens were stored dry on silica gel before…
Partner study description Mosquitoes were collected by landing catches in the capital city Libreville (0.384, 9.455) in December 2000 (1), an urban and polluted site. Malaria is endemic throughout the year. Specimens were…
Partner study description Samples were collected in Twifo Praso (5.609, -1.549), a peri-urban community located in semi-deciduous forest in the Central Region of Ghana. It is an extensive agricultural area characterised by small-scale…
Partner study description Collections were made from four different study sites around the border between Guinea and Mali. From Mali; Takan (11.47, -8.33) and Toumani Oulena (10.83, -7.81) are both small villages in…
Partner study description Guinea Bissau samples were collected from three sites in October 2010 by indoor CDC light traps. Safim (11.957, -15.649) and Antula (11.891, -15.582), from a south-western coastal region, characterised mainly…
Partner study desrcription Kenyan specimens were obtained from villages located in Kilifi County near the Kenyan coast between 2000 and 2014. All Anopheles mosquito sampling was conducted indoors using CDC light traps which…
Partner study description Specimens were obtained from villages within the catchment of the Majete Malaria Project, Chikhwawa District, Malawi (-15.933, 34.755) (1). Mosquitoes were collected indoors and outdoors by Suna light trap from…
Partner study description Collections were made in four villages in the Koulikoro region; Tieneguebougou (12.810, -8.080) approximately 20km north of Bamako, and Kababougou (12.890, -8.150), Ouassorola (12.900, -8.160), Sogolombougou (12.880, -8.140), approximately 30km…
Partner study description Collections of indoor resting adults were made by spray catch from seven villages in the southern part of Mali in August-September 2004: Banambani (12.800, -8.050), Bancoumana (12.200, -8.200), Douna (13.210,…
Partner study description Collections were taken from multiple sites on the island of Mayotte. Samples were collected as larvae during March-April 2011 in temporary pools by dipping. Sites included Mtsanga Charifou (-12.991, 45.156)…
Partner study description Mosquito samples were collected in Furvela (-23.716, 35.299), Mozambique, by CDC light traps between December 2003 and April 2004. Specimens were stored on silica gel and DNA was extracted according…
Partner study description Tanzanian samples were collected from four distinct locations. Moshi samples came from lower Mabogini (-3.400, 37.350), rice fields near lower Moshi on the southern slope of Mount Kilimanjaro, a region…
Partner study description Indoor resting female mosquitoes were collected by pyrethrum spray catch from four hamlets around Njabakunda (13.55, -15.90), North Bank Region, The Gambia between August and October 2011. The four hamlets…
Partner study description Specimens were collected along the Gambia River from the western coastal region of The Gambia (1), in August 2006. An. gambiae and An. coluzzii specimens were identified to species following…
Partner study description Adult mosquitoes were collected at Wali Kunda in the rural, central river region of The Gambia (13.567, -14.917). The area is 180km from the sea, on the south bank of…
Partner study description Specimens were obtained from two locations in Uganda: Nagongera, 30km to the North of Lake Victoria near the border with Kenya, and Kihihi, in the very South-West of the country.…
Publications
- Speciation within the Anopheles gambiae complex: high-throughput whole genome sequencing reveals evidence of a putative new cryptic taxon in 'far-west' Africa
Caputo et al.Commun Biol, 2024; 7 1115- Variation in spatial population structure in the Anopheles gambiae species complex
McCann et al.bioRxiv, 2024;- A metagenomic analysis of the phase 2 Anopheles gambiae 1000 genomes dataset reveals a wide diversity of cobionts associated with field collected mosquitoes
Pastusiak et al.Communications Biology, 2024;- Evaluating evidence for co-geography in the Anopheles–Plasmodium host-parasite system
Rehmann et al.G3 Genes Genomes Genetics, 2024; 14 (3)- Parallel evolution in mosquito vectors – a duplicated esterase locus is associated with resistance to pirimiphos-methyl in An. gambiae
Nagi et al.bioRxiv, 2024;- Whole-genome sequencing of major malaria vectors revealed an evolution of new insecticide resistance variants in a longitudinal study in Burkina Faso
Kientega et al.bioRxiv, 2023;- Dispersal inference from population genetic variation using a convolutional neural network
Smith et al.Genetics, 2023; 224(2)- Single nucleotide polymorphism (SNP) in the doublesex (dsx) gene splice sites and relevance for its alternative splicing in the malaria vector Anopheles gambiae
Djihinto et al.Wellcome Open Research, 2023; 7(31)- RNA-Seq-Pop: Exploiting the sequence in RNA-Seq - a Snakemake workflow reveals patterns of insecticide resistance in the malaria vector Anopheles gambiae
Nagi et al.Molecular Ecology Resources, 2023; 23(4) 946-961- Analysis of the Genetic Variation of the Fruitless Gene within the Anopheles gambiae (Diptera: Culicidae) Complex Populations in Africa
Kientega et al.Insects, 2022; 13(11) 1048- Genetic variation at the Cyp6m2 putative insecticide resistance locus in Anopheles gambiae and Anopheles coluzzii
Wagah et al.Malaria Journal, 2021; 20 234- Contemporary Ne estimation using temporally spaced data with linked loci
Hui et al.Molecular Ecology Resources, 2021;- Discovery of Ongoing Selective Sweeps within Anopheles Mosquito Populations Using Deep Learning
Xue et al.Mol Biol Evol, 2021; 38 1168-1183- The genetic architecture of target-site resistance to pyrethroid insecticides in the African malaria vectors Anopheles gambiae and Anopheles coluzzii
Clarkson et al.Molecular Ecology, 2021;- Novel genotyping approaches to easily detect genomic admixture between the major Afrotropical malaria vector species, Anopheles coluzzii and An. gambiae
Caputo et al.Molecular Ecology Resources, 2021;- Resistance to pirimiphos-methyl in West African Anopheles is spreading via duplication and introgression of the Ace1 locus.
Grau-Bové et al.PLoS Genetics, 2021; 17- Evolution of the Insecticide Target Rdl in African Anopheles Is Driven by Interspecific and Interkaryotypic Introgression
Grau-Bové et al.Mol Biol Evol, 2020; 1 37- Genome variation and population structure among 1142 mosquitoes of the African malaria vector species Anopheles gambiae and Anopheles coluzzii
The Anopheles gambiae 1000 Genomes ConsortiumGenome Research, 2020; 30 1533-1546- Whole-genome sequencing reveals high complexity of copy number variation at insecticide resistance loci in malaria mosquitoes
Lucas et al.Genome Research, 2019;- Robust Estimation of Recent Effective Population Size from Number of Independent Origins in Soft Sweeps
Khatri and BurtMolecular Biology and Evolution, 2019;- Genetic diversity of the African malaria vector Anopheles gambiae
The Anopheles gambiae 1000 Genomes ConsortiumNature, 2017;- Panoptes: web-based exploration of large scale genome variation data
Vauterin et al.Bioinformatics, 2017;- Adaptive introgression between Anopheles sibling species eliminates a major genomic island but not reproductive isolation
Clarkson et al.Nature Communications, 2014; 5 4248
765 mosquitoes
from
8 countries
Project contact
People
The Ag1000G Consortium involves researchers from several institutions around the world, including experts in Anopheles genomics, population genetics, epidemiology, and vector biology. The Ag1000G Consortium comprises members of two working groups, as well as affiliates involved in data production and analysis.
Partner Working Group
The Partner Working Group (PWG) is comprised of the investigators contributing samples or other significant resources / strategic guidance to the Project.
- Alessandra della Torre
- Andrew Kern
- Beniamino Caputo
- Dr Lucas Amenga-Etego
- Bilali Kabula
- Bradley White
- Prof Charles Godfray
- Constant Edi
- Dr Craig Wilding
- Caterina A Fanello
- Dr Daniel Neafsey
- Professor Milijaona Randrianarivelojosia
- Daniel Schrider
- Prof David Conway
- Dr David Weetman
- Prof Francois Nosten
- Diego Ayala
- Dominic Kwiatkowski
- Dr Sarah Auburn
- Prof Igor Sharakhov
- Dr Janet Midega
- Colin Sutherland
- Dr Jiannong (John) Xu
- Dr João Pinto
- John Essandoh
- Johnson Matowo
- Dr Ken Vernick
- Dr Luc S. Djogbenou
- Mamadou Coulibaly
- Dr Mara Lawniczak
- Prof Martin Donnelly
- Prof Matthew Hahn
- Dr Michaël Fontaine
- Dr Michelle Riehle
- Prof Nora Besansky
- Dr Omar Cornejo
- Robert McCann
- Dr Sam O’Loughlin
- Vincent Robert
Data Analysis Working Group
The Data Analysis Group (DAG) is undertaking core project analyses and includes members of both the PWG and PMG, alongside additional members of the Ag1000G Consortium.
- Alistair Miles
- Chris Clarkson
- Daniel Schrider
- Dominic Kwiatkowski
- Dr Frederic Labbe
- Giordano Bottà
- Dr Janet Midega
- Jeffrey Adrion
- Dr Jiannong (John) Xu
- Joel Nelson
- Dr Mara Lawniczak
- Prof Martin Donnelly
- Prof Matthew Hahn
- Dr Michaël Fontaine
- Dr Nick Harding
- Dr Richard Wang
- Dr Sam O’Loughlin
- Dr Scott T Small
- Abdelrahim Osman Mohamed
- Dr Seth Redmond
- Dr Tiago Antão
- Cody Champion
- Alexander Xue
- CJ Battey
Updates
6 Oct 2015
This data is subject to