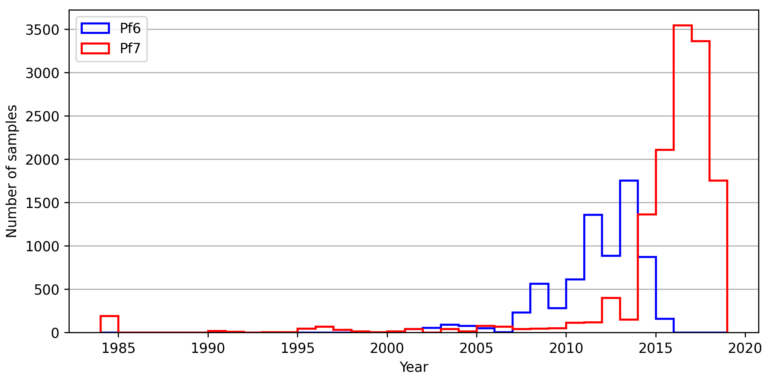
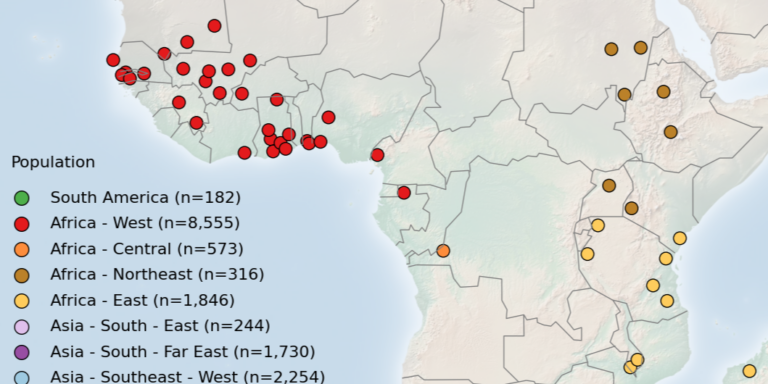
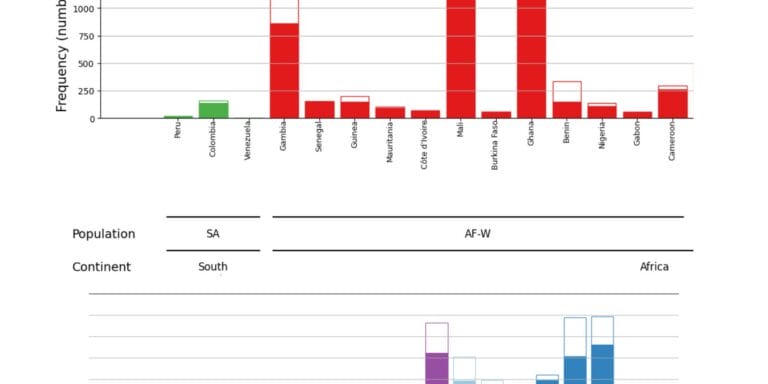
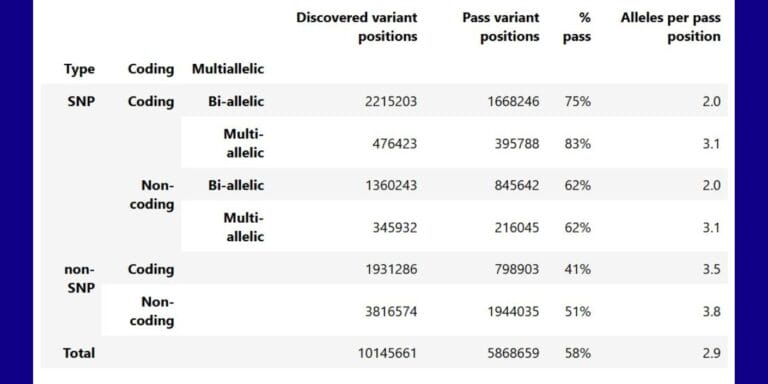
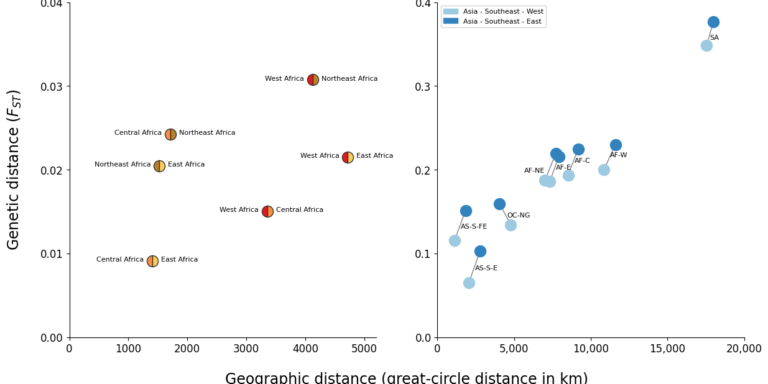
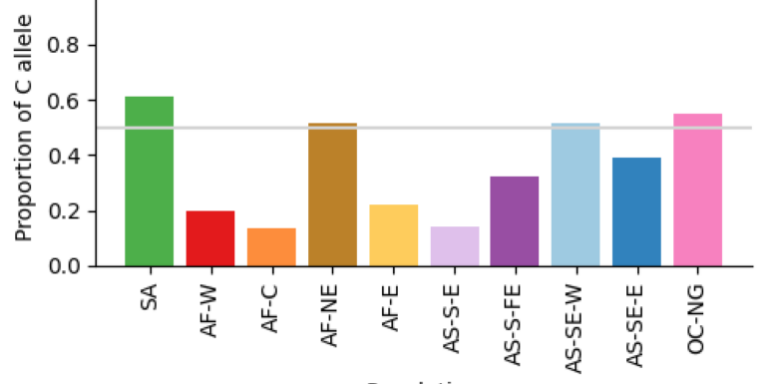
Geographical variation drives adaptive equilibrium of the P. falciparum sickle-associated mutations
Andre Python, Annie J. Forster, Peter Todd, Karamoko Niaré, Melissa D. Conrad, Lucas N. Amenga-Etego, Deus S. Ishengoma, Jianjun Lian, Baoli Liu, Yucheng Yan, Jingmin Liang, Muhammad M. Khan, Junkan Zheng, Jeffrey A. Bailey, David J. Conway, Alexander J. Mentzer, Alfred Amambua-Ngwa, Ellen M. Leffler, Thomas N. Williams, eGavin Band
bioRxiv 2025
DOI: 10.1101/2025.08.31.672853
Identification of Novel Gene Cluster Potentially Associated with Insecticide Resistance in Anopheles gambiae s.l.
Hyacinthe Dipina Ki, Mahamadi Kientega, Sabéré O. G. Yemien, Hamidou Maiga, Nouhoun Traoré, Koama Bayili, Moussa Namountougou, Abdoulaye Diabaté
Genes 2025;16(9), 1018
DOI: 10.3390/genes16091018
Global-scale population genetic analysis of Plasmodium falciparum identifies country- and region-specific patterns of malaria parasite adaptation
Nina Billows, Jamille G. Dombrowski, Joseph Thorpe, Leen Vanheer, Sophie Moss, Jesse Gitaka, Colin Sutherland, Claudio R. F. Marinho,Nguyen Ngoc TH, Nguyen Binh TH, Nguyen Thieu Q, Susana Campino, Taane Clark
Research Square 2025
DOI: 10.21203/rs.3.rs-7160640/v1
Worldwide genetic diversity of Plasmodium vivax Pv47 is consistent with natural selection by anopheline mosquitoes
Alvaro Molina-Cruz, Lilia Gonzalez-Ceron, Ankit Dwivedi, Tran Zen B. Torres, Nadia Raytselis, Micah Young, Nitin Kamath, Colton McNinch, Xinzhuan Su, Anthony Ford, Marcelo U. Ferreira, Myriam Arévalo-Herrera, Sócrates Herrera, Eugenia Lo, Joana C. Silva & Carolina Barillas-Mury
Nature Communications 2025;16, 7363
DOI: 10.1038/s41467-025-62680-3
A novel locus associated with decreased susceptibility of Plasmodium falciparum to lumefantrine and dihydroartemisinin has emerged and spread in Uganda
Karamoko Niaré, Bersabeh Tafesse, Mayland Treat, Jacob M. Sadler, Martin Okitwi, Stephen Orena, Victor Asua, Oriana Kreutzfeld, Jenny Legac, Samuel L. Nsobya, Adoke Yeka, Philip J. Rosenthal, Jonathan J. Juliano, Jeffrey A Bailey, Melissa D. Conrad
bioRxiv 2025
DOI: 10.1101/2025.07.30.667738
Computational Prioritization of T Cell Epitopes to Overcome HLA Restriction and Antigenic Diversity in Plasmodium falciparum
Alexander J. Laurenson, Brian G. Pierce, Shannon Takala-Harrison, Matthew B. Laurens
bioRxiv 2025
DOI: 10.1101/2025.07.14.664425
Molecular Characterisation of Plasmodium falciparum in Children with Uncomplicated Malaria in Homa-Bay, Kenya; Two Decades Post-Adoption of Artemisinin-Based Combination Therapies
Florence T. Akinyi, Joyce B Oluokun, Kabir Gorden, John Openibo, Clemence D. Gouton, Oloo Edwin O, Christian T. Happi, Vera Mitsser, Onikepe Folarin,
Research Square 2025
DOI: 10.21203/rs.3.rs-6897552/v1
DAPCy: a Python package for the discriminant analysis of principal components method for population genetic analyses
Alejandro Correa Rojo, Pieter Moris, Hanne Meuwissen, Pieter Monsieurs, Dirk Valkenborg
Bioinformatics Advances 2025
DOI: 10.1093/bioadv/vbaf143
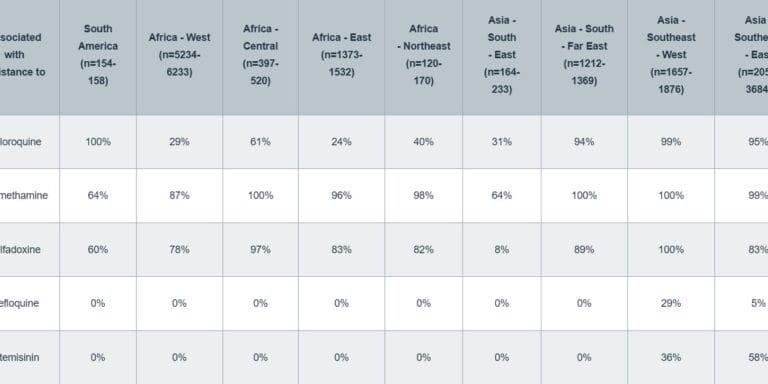
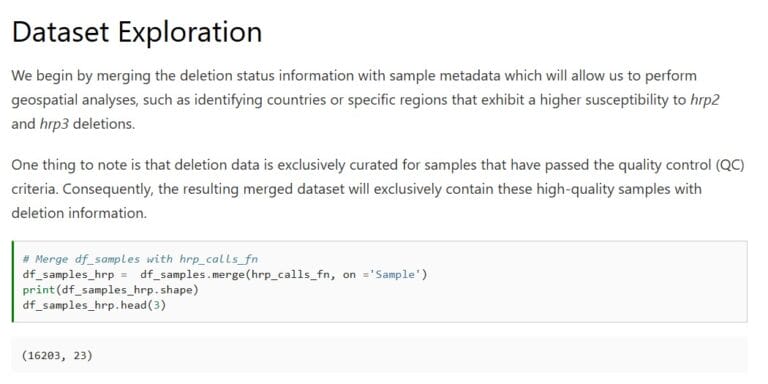
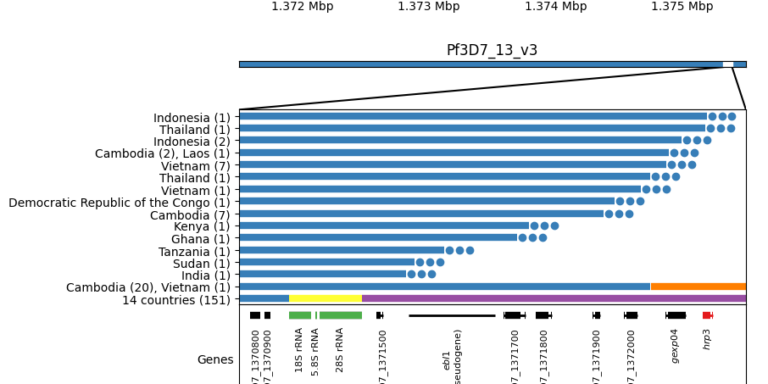
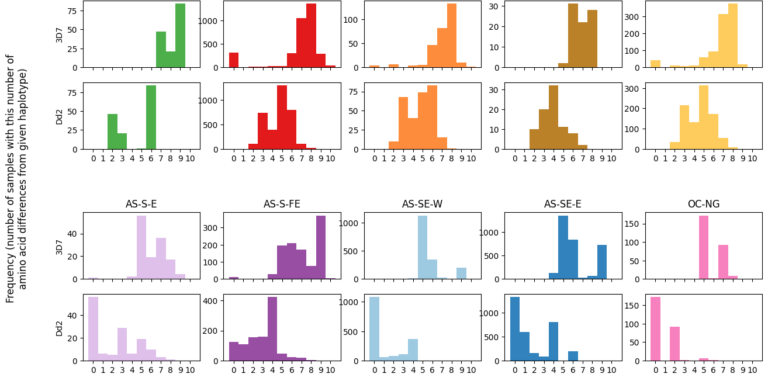
Cryptic absence and genetic variation of Plasmodium falciparum PfHRP2 and PfHRP3 from isolates in Papua, Indonesia
Agatha Mia Puspitasari, Edwin Sutanto, Khilal Syauqi, Ristya Amalia, Fahira Ainun Nisa, Leily Trianty, Elizabeth Sidhartha, Enny Kenangalem, Jeanne Rini Poespoprodjo, Rukhsana Ahmed, Ewurama Owusu, Lise Carlier, Kevin K. A. Tetteh, Qin Cheng & Rintis Noviyanti
Scientific Reports 2025;15, 19511
DOI: 10.1038/s41598-025-02834-x
hmmibd-rs: An enhanced hmmIBD implementation for parallelizable identity-by-descent detection from large-scale Plasmodium genomic data
Bing Guo, Stephen F. Schaffner, Aimee R. Taylor, Timothy D. O’Connor, Shannon Takala-Harrison
Research Square 2025
DOI: 10.21203/rs.3.rs-7004070/v1
Selection of Plasmodium falciparum kelch13 mutations in Uganda in comparison with southeast Asia: a modelling study
Cecile P G Meier-Scherling, Oliver J Watson, Victor Asua, Isaac Ghinai, Thomas Katairo, Shreeya Garg, Melissa D Conrad, Prof Philip J Rosenthal, Lucy C Okell, Jeffrey A Bailey
The Lancet Microbe 2025;6;5
DOI: 10.1016/j.lanmic.2024.101027
Detection of novel Plasmodium falciparum haplotypes under treatment pressure in paediatric severe malaria
Balotin Fogang, Emilie Guillochon, Claire Kamaliddin, Gino Agbota, Sem Ezinmegnon, Maroufou Jules Alao, Philippe Deloron, Gwladys Bertin and Antoine Claessens
Microbiology Society 2025;11;5
DOI: 10.1099/mgen.0.001386
PvGAP: Development of a globally-applicable, highly-multiplexed microhaplotype amplicon panel for Plasmodium vivax
Alfred Hubbard, Edwin Solares, Lauren Bradley, Brook Jeang, Delenasaw Yewhalaw, Daniel Janies, Eugenia Lo, Guiyun Yan, Elizabeth Hemming-Schroeder
medRxiv 2025
DOI: 10.1101/2025.04.30.25326751
Pregnant women as a sentinel population for genomic surveillance of malaria in the Democratic Republic of the Congo: a population-based study
Marie Onyamboko, Varanya Wasakul, Sarah Benie Bakomba, Daddy Kalala Kayembe, Bejos Kifakiou Nzambiwishe, Pascal Epe Ekombolo, Benjamen Basara Badjanga, Jean-Robert Moke Maindombe, Jephte Ndundu Ngavuka, Brunette Nsunda Lwadi, Eleanor Drury, Cristina Ariani, Sonia Goncalves, Vanapol Chamsukhee, Naomi Waithira, Tess D Verschuuren, Sue J Lee, Olivo Miotto, Caterina Fanello
The Lancet Global Health 2025;13;3
DOI: 10.1016/S2214-109X(24)00497-2
Globally prevalent Kelch13 mutations increase partial artemisinin resistance and fitness in Bangladeshi Plasmodium falciparum parasites
Maisha Khair Nima, Nirjhar Bhattacharyya, Jungyoon Park, Douglas Shoue, Ching Swe Phru, Saiful Arefeen Sazed, Sudhir Kumar, Mohammad Shafiul Alam, Michael Ferdig, Angana Mukherjee
bioRxiv 2025
DOI: 10.1101/2025.04.25.650672
In vitro evaluation of multi-protein chimeric antigens in effectively clearing the blood stage of Plasmodium falciparum
Bhagyashree Deshmukh, Dhruv Khatri, Sanjay Kumar Kochar , Chaitanya Athale, Krishanpal Karmodiya
Vaccine 2025;54
DOI: 10.1016/j.vaccine.2025.126952
Disparate co-evolution and prevalence of sulfadoxine and pyrimethamine resistance alleles and haplotypes at dhfr and dhps genes across Africa
Nina F. D. White, Georgia Whitton, Varanya Wasakul, Lucas Amenga-Etego, Antoine Dara, Voahangy Andrianaranjaka, Milijaona Randrianarivelojosia, Olivo Miotto, Umberto D’Alessandro, Abdoulaye Djimdé, Cristina V. Ariani, Richard D. Pearson & Alfred Amambua-Ngwa
Nature Scientific Reports 2025;15;13222
DOI: 10.1038/s41598-025-98035-7
Plasmodium falciparum Subtilisin-Like Domain Containing Protein ((PfSDP) PF3D7_1105800), a Cross-Stage Antigen, Elicits Short-Lived Antibody Response Following Natural Infection with Plasmodium falciparum
Jonas Arnaud Kengne-Ouafo, Collins M Morang'a, Nancy K Nyakoe, Daniel Dosoo, Richmond Tackie, Joe Mutungi, Saikou Y Bah, Lucas Amenga-Etego, Britta C. Urban, Gordon A. Awandare, Yaw Aniweh, Bismarck Dinko
Preprints.org 2025
DOI: 10.20944/preprints202504.0407.v
A monoclonal antibody selectively recognizing PfEMP1 proteins associated with cerebral malaria
Nanna Dalgaard, Rebecca W Olsen, Yvonne Adams, Blanca L Mendez, Sofie Amalie Gehrcke Bisholm, Jonas J Rudbaek, Azizath Mousiliou, Melanie R Walker, Lars Hviid, Rachida Tahar, Nicaise T Ndam2 Lea Barfod1 Email Anja R Jensen1
Research Square 2025
DOI: 10.21203/rs.3.rs-6252569/v1
An expanded method for malaria parasite genetic surveillance using targeted nanopore sequencing
William L. Hamilton, Lucas N. Amenga-Etego, Alexandria J. R. Harrott, Richard D. Pearson, Cristina V. Ariani, Collins M. Morang'a, Mona-Liza Sakyi, Ahmed Osumanu, Enock K. Amoako, Fagdéba David Bara, Yaw Aniweh, Gordon A. Awandare, Myra Hosmillo , Kess Rowe , Ian Goodfellow , Francis Zeukeng 2:30
veRixiv 2025
DOI: 10.12688/verixiv.630.1
Highly multiplex molecular inversion probe panel in Plasmodium falciparum targeting common SNPs approximates whole genome sequencing assessments for selection and relatedness
Karamoko Niaré, Rebecca Crudale, Abebe A. Fola, Neeva Wernsman Young, Victor Asua, View ORCID ProfileMelissa Conrad, Pierre Gashema, Anita Ghansah, Stan Hangi, Deus S. Ishengoma, Jean-Baptiste Mazarati, Ayalew Jejaw Zeleke, Philip J. Rosenthal, View ORCID ProfileAbdoulaye A. Djimdé, Jonathan J. Juliano, Jeffrey A Bailey
medRxiv 2025
DOI: 10.1101/2025.03.07.25323597
New insights on malaria parasite adaptation yielded by samples archived for more than 50 years
Alfred Amambua-Ngwa, Mouhamadou Fadel Diop, Christopher J. Drakeley, Umberto d’Alessandro, Dominic P. Kwiatkowski, David J. Conway
bioRxiv 2025
DOI: 10.1101/2025.03.11.642566
Liver stage P. falciparum antigens highly targeted by CD4+ T cells in malaria-exposed Ugandan children
Gonzalo R. Acevedo, Sophie S. Samiee, Mikias Ilala, Justine Levan, Meagan E. Olive, Riana D. Hunter, Mary Prahl, Raja Rajalingam, John Rek, Grant Dorsey, Margaret E. Feeney
PLOS Pathogens 2025;21;2
DOI: 10.1371/journal.ppat.1012943
Heterogeneous constraint and adaptation across the malaria parasite life cycle
Sarah A. Perkins, Daniel E. Neafsey, Angela M. Early
bioRxiv 2025
DOI: 10.1101/2025.02.11.636054
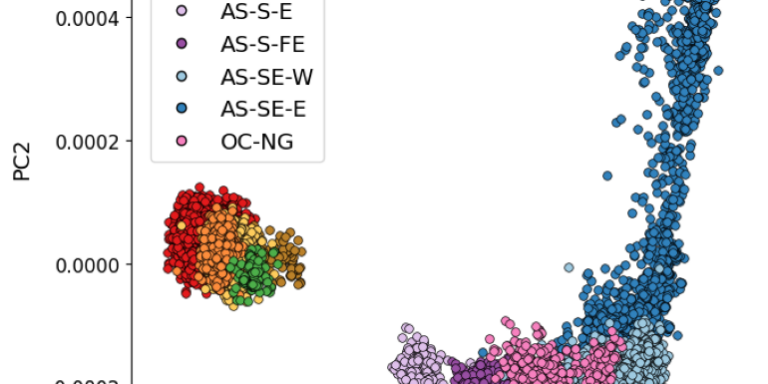
Genomic exploration of the journey of Plasmodium vivax in Latin America
Margaux J. M. Lefebvre, Fanny Degrugillier, Céline Arnathau, Gustavo A. Fontecha, Oscar Noya, Sandrine Houzé, Carlo Severini, Bruno Pradines, Antoine Berry, Jean-François Trape, Fabian E. Sáenz, Franck Prugnolle, Michael C. Fontaine, Virginie Rougeron
PLOS Pathogens 2025;21:1
DOI: 10.1371/journal.ppat.1012811
Role of novel mutations in food vacuole transporters beyond K13-mediated artemisinin resistance in Plasmodium falciparum
Iqbal Taliy Junaid, Ashutosh Panda, Arunaditya Deshmukh, Rahila Sardar, Monika Narwal, Prakhar Agrawal, Neha Prakash, Asif Akhtar, Amit Kumar Dey, Suneet Shekhar Singh, Saptarshi Mridha, Jigneshkumar Mochi, Sadaf Parveen, Mohit Kumar, Rashi Nagar, Naseem Gaur, Dinesh Gupta, Asif Mohmmed, Inderjeet Kaur, Krishanpal Karmodiya, Pawan Malhotra
Research Square 2025
DOI: 10.21203/rs.3.rs-5749310/v1
Plasmodium falciparum transcription factor AP2-06B is mutated at high frequency in Southeast Asia but does not associate with drug resistance
Qiyang Shi, Changhong Wang, Wenluan Yang, Xiaoqin Ma. Jianxia Tang, Jiayao Zhang, Guoding Zhu, Yinlong Wang, Yaobao Liu, Xiaoqin He
Front. Cell. Infect. Microbiol. 2025;14
DOI: 10.3389/fcimb.2024.1521152
Protective antibodies target cryptic epitope unmasked by cleavage of malaria sporozoite protein
Cherrelle Dacon, Re’em Moskovitz, Kristian Swearingen, Lais Da Silva Pereira, Yevel Flores-Garcia, Maya Aleshnick, Sachie Kanatani, Barbara Flynn, Alvaro Molina-Cruz, Kurt Wollenberg, Maria Traver, Payton Kirtley, Lauren Purser, Marlon Dillon, Brian Bonilla, Adriano Franco, Samantha Petros, Jake Kritzberg, Courtney Tucker, Gonzalo Gonzalez Paez, Priya Gupta, Melanie J. Shears, Joseph Pazzi, Joshua M. Edgar, Andy A. Teng, Arnel Belmonte, Kyosuke Oda, Safiatou Doumbo, Ludmila Krymskaya, Jeff Skinner, Shanping Li, Suman Ghosal, Kassoum Kayentao, Aissata Ongoiba, Ashley Vaughan, Joseph J. Campo, Boubacar Traore, Carolina Barillas-Mury, Wathsala Wijayalath, Azza Idris, Peter D. Crompton, Photini Sinnis, Brandon K. Wilder, Fidel Zavala, Robert A. Seder, Ian A. Wilson, and Joshua Tan
Science 2025;387(6729)
DOI: 10.1126/science.adr0510
Regular Plasmodium falciparum importation onto Bioko Island, Equatorial Guinea, hampers malaria elimination from the island
Thomas C. Stabler, Ankit Dwivedi, Bing Guo, Biraj Shrestha, Sudhaunshu Joshi, Matilde Riloha Rivas , Olivier Tresor Donfack , Carlos A. Guerra , Guillermo A. García , Claudia Daubenberger, Joana C. Silva
bioRxiv 2024
DOI: 10.1101/2024.12.19.629489
Geo-classification of drug-resistant travel-associated Plasmodium falciparum using Pfs47 and Pfcpmp gene sequences (USA, 2018–2021)
Edwin Pierre-Louis, Julia Kelley, Dhruviben Patel, Christina Carlson, Eldin Talundzic, David Jacobson, Joel Leonard, Nicholas Barratt
Antimicrobial Agents and Chemotherapy 2024;68:12
DOI: 10.1128/aac.01203-24
Pf-HaploAtlas: an interactive web app for spatiotemporal analysis of Plasmodium falciparum genes
Chiyun Lee, Eyyüb S Ünlü, Nina F D White, Jacob Almagro-Garcia, Cristina V Ariani, Richard D Pearson
Bioinformatics 2024;40:11
DOI: 10.1093/bioinformatics/btae673
Identification of complex Plasmodium falciparum genetic backgrounds circulating in Africa: a multicountry genomic epidemiology analysis
Olivo Miotto, Alfred Amambua-Ngwa, Lucas N Amenga-Etego, Muzamil M Abdel Hamid, Ishag Adam, Enoch Aninagyei, Tobias Apinjoh, Prof Gordon A Awandare, Philip Bejon, Gwladys I Bertin, Marielle Bouyou-Akotet, Antoine Claessens, David J Conway, Prof Umberto D'Alessandro, Prof Mahamadou Diakite, Abdoulaye Djimdé, Arjen M Dondorp, Patrick Duffy, Rick M Fairhurst, Caterina I Fanello, Anita Ghansah, Deus S Ishengoma, Mara Lawniczak, Oumou Maïga-Ascofaré, Sarah Auburn, Anna Rosanas-Urgell, Varanya Wasakul, Nina F D White, Alexandria Harrott, Jacob Almagro-Garcia, Richard D Pearson, Sonia Goncalves, Cristina Ariani, Prof Zbynek Bozdech, William L Hamilton, Victoria Simpson, Prof Dominic P Kwiatkowski.
The Lancet Microbe 2024;5:12
DOI: 100941 10.1016/j.lanmic.2024.07.004
Genetic Diversity of the Plasmodium falciparum Reticulocyte Binding protein Homologue-5 which is a potential Malaria Vaccine Candidate: Baseline data from areas of varying malaria endemicity in Mainland Tanzania
Angelina J. Kisambale, Beatus M. Lyimo, Dativa Pereus, Salehe S. Mandai, Catherine Bakari, Gervas A. Chacha, Ruth B. Mbwambo, Ramadhan Moshi, Daniel A. Petro, Daniel P. Challe, Misago D. Seth, Rashid A. Madebe, Rule Budodo, Sijenunu Aaron, Daniel Mbwambo, Abdallah Lusasi, Stella Kajange, Samwel Lazaro, Ntuli Kapologwe, Celine I. Mandara, Deus S. Ishengoma
medRxiv 2024
DOI: 10.1101/2024.09.20.24314052
Characterisation of the erythrocyte invasion phenotype of FCB-2: A South American P. falciparum reference strain
Monica Ararat-Sarria, Hernando Curtidor and Manuel Alfonso Patarroyo
Acta Tropica 2024;260;107379
DOI: 10.1016/j.actatropica.2024.107379
ResMAP—a saturation mutagenesis platform enabling parallel profiling of target-specific resistance-conferring mutations in Plasmodium
Richard J. Wall, Stuart A. MacGowan, Irene Hallyburton, Aisha J. Syed, Sowmya Ajay Castro, Gourav Dey, Rachel Milne, Stephen Patterson, Jody Phelan, Natalie Wiedemar, Susan Wyllie
mBio 2024;e0170824
DOI: 10.1128/mbio.01708-24
Detection of novel Plasmodium falciparum haplotypes under treatment pressure in pediatric severe malaria
Balotin Fogang, Emilie Guillochon, Claire Kamaliddin, Gino Agbota, Sem Ezinmegnon, Maroufou Jules Alao, Philippe Deloron, Gwladys Bertin, Antoine Claessens
medRxiv 2024
DOI: 10.1101/2024.08.20.24312296
Household clustering and seasonal genetic variation of Plasmodium falciparum at the community-level in The Gambia
Marc-Antoine Guery, Sukai Ceesay, Sainabou Drammeh, Fatou K Jaiteh, Umberto d’Alessandro, Teun Bousema, David J Conway, Antoine Claessens
medRxiv 2024
DOI: 10.1101/2024.08.05.24311344
Diversity and selection analyses identify transmission-blocking antigens as the optimal vaccine candidates in Plasmodium falciparum
Ilinca I Ciubotariu, Bradley K Broyles, Shaojun Xie, Jyothi Thimmapuram, Mulenga C Mwenda, Brenda Mambwe, Conceptor Mulube, Japhet Matoba, Jessica L Schue, William J Moss, Daniel J Bridges, Qixin He, Giovanna Carpi
eBioMedicine 2024;106;105227
DOI: 10.1016/j.ebiom.2024.105227
Benchmarking and Optimization of Methods for the Detection of Identity-By-Descent in High-Recombining Plasmodium falciparum Genomes
Bing Guo, Shannon Takala-Harrison, Timothy D. O’Connor
bioRxiv 2024
DOI: 10.1101/2024.05.04.592538
A Plasmodium falciparum genetic cross reveals the contributions of pfcrt and plasmepsin II/III to piperaquine drug resistance
John Kane, Xue Li, Sudhir Kumar, Katrina A. Button-Simons, Katelyn M. Vendrely Brenneman, Haley Dahlhoff, Mackenzie A. C. Sievert, Lisa A. Checkley, Douglas A. Shoue, Puspendra P. Singh, Biley A. Abatiyow, Meseret T. Haile, Shalini Nair, Ann Reyes, Rupam Tripura, Thomas J. Peto, Dysoley Lek, Angana Mukherjee, Stefan H. I. Kappe, Mehul Dhorda, Standwell C. Nkhoma, Ian H. Cheeseman, Ashley M. Vaughan, Timothy J. C. Anderson, Michael T. Ferdig
mBio 2024;15;e00805-24
DOI: 10.1128/mbio.00805-24
Ancient Plasmodium genomes shed light on the history of human malaria
Megan Michel, Eirini Skourtanioti, Federica Pierini, Evelyn K. Guevara, Angela Mötsch, Arthur Kocher, Rodrigo Barquera, Raffaela A. Bianco, Selina Carlhoff, Lorenza Coppola Bove, Suzanne Freilich, Karen Giffin, Taylor Hermes, Alina Hiß, Florian Knolle, Elizabeth A. Nelson, Gunnar U. Neumann, Luka Papac, Sandra Penske, Adam B. Rohrlach, Nada Salem, Lena Semerau, Vanessa Villalba-Mouco, Isabelle Abadie, Mark Aldenderfer, Jessica F. Beckett, Matthew Brown, Franco G. R. Campus, Tsang Chenghwa, María Cruz Berrocal, Ladislav Damašek, Kellie Sara Duffett Carlson, Raphaël Durand, Michal Ernée, Cristinel Fântăneanu, Hannah Frenzel, Gabriel García Atiénzar, Sonia Guillén, Ellen Hsieh, Maciej Karwowski, David Kelvin, Nikki Kelvin, Alexander Khokhlov, Rebecca L. Kinaston, Arkadii Korolev, Kim-Louise Krettek, Mario Küßner, Luca Lai, Cory Look, Kerttu Majander, Kirsten Mandl, Vittorio Mazzarello, Michael McCormick, Patxuka de Miguel Ibáñez, Reg Murphy, Rita E. Németh, Kerkko Nordqvist, Friederike Novotny, Martin Obenaus, Lauro Olmo-Enciso, Päivi Onkamo, Jörg Orschiedt, Valerii Patrushev, Sanni Peltola, Alejandro Romero, Salvatore Rubino, Antti Sajantila, Domingo C. Salazar-García, Elena Serrano, Shapulat Shaydullaev, Emanuela Sias, Mario Šlaus, Ladislav Stančo, Treena Swanston, Maria Teschler-Nicola, Frederique Valentin, Katrien Van de Vijver, Tamara L. Varney, Alfonso Vigil-Escalera Guirado, Christopher K. Waters, Estella Weiss-Krejci, Eduard Winter, Thiseas C. Lamnidis, Kay Prüfer, Kathrin Nägele, Maria Spyrou, Stephan Schiffels, Philipp W. Stockhammer, Wolfgang Haak, Cosimo Posth, Christina Warinner, Kirsten I. Bos, Alexander Herbig & Johannes Krause
Nature 2024;631;125–133
DOI: 10.1038/s41586-024-07546-2
Genetic surveillance of Plasmodium falciparum reveals rapid population changes following first-line treatment policy revisions in the Greater Mekong Subregion
Tess D Verschuuren, Varanya Wasakul, Nguyen Thuy-Nhien, Ethan Booth, Huynh Hong Quang, Ngo Duc Thang, Keobouphaphone Chindavongsa, Siv Sovannaroth, Virasak Banouvong, Viengphone Sengsavath, Mayfong Mayxay, Nguyen Thi Kim Tuyen, Vo Ngoc Lam Phuong, Pham Duc Trung, Sónia Gonçalves, Soun Chen, Sonexay Phalivong, Saiamphone Xayvanghang, Supaporn Mahaphontrakoon, Richard D Pearson, Paul N Newton, Richard J Maude, Elizabeth A Ashley, Cristina V Ariani, Victoria J Simpson, Nicholas P Day, Arjen M Dondorp, Olivo Miotto
medRxiv 2024
DOI: 10.1101/2024.06.06.24308535
Molecular markers of artemisinin resistance during falciparum malaria elimination in Eastern Myanmar
Aung Myint Thu, Aung Pyae Phyo, Chanapat Pateekhum, Jade D. Rae, Jordi Landier, Daniel M. Parker, Gilles Delmas, Wanitda Watthanaworawit, Alistair R. D. McLean, Ann Arya, Ann Reyes, Xue Li, Olivo Miotto, Kyaw Soe, Elizabeth A. Ashley, Arjen Dondorp, Nicholas J. White, Nicholas P. Day, Tim J. C. Anderson, Mallika Imwong, Francois Nosten & Frank Smithuis
Malaria Journal 2024;23(138)
DOI: 10.1186/s12936-024-04955-6
Genetic polymorphism and evidence of signatures of selection in the Plasmodium falciparum circumsporozoite protein gene in Tanzanian regions with different malaria endemicity
Beatus M. Lyimo, Catherine Bakari, Zachary R. Popkin-Hall, David J. Giesbrecht, Misago D. Seth, Dativa Pereus, Zulfa I. Shabani, Ramadhan Moshi, Ruth Boniface, Celine I. Mandara, Rashid Madebe, Jonathan J. Juliano, Jeffrey A. Bailey & Deus S. Ishengoma
Malaria Journal 2024;23(1);139
DOI: 10.1186/s12936-024-04974-3
Sustained clinical benefit of malaria chemoprevention with sulfadoxine-pyrimethamine (SP) in pregnant women in a region with high SP resistance markers
Glória Matambisso, Nanna Brokhattingen, Sónia Maculuve, Pau Cístero, Henriques Mbeve, Anna Escoda, Gizela Bambo, Boaventura Cuna, Cardoso Melembe, Nelo Ndimande, Kevin K.A. Tetteh, Chris Drakeley, Benoit Gamain, Chetan Chitnis, Virander Chauhan, Llorenç Quintó, Eusébio Macete, Alfredo Mayor
Journal of Infection 2024;88;(5)
DOI: 10.1016/j.jinf.2024.106144
Strong positive selection biases identity-by-descent-based inferences of recent demography and population structure in Plasmodium falciparum
Bing Guo, Victor Borda, Roland Laboulaye, Michele D. Spring, Mariusz Wojnarski, Brian A. Vesely, Joana C. Silva, Norman C. Waters, Timothy D. O’Connor & Shannon Takala-Harrison
Nat Commun 2024;15;2499
DOI: 10.1038/s41467-024-46659-0
Targeted amplicon deep sequencing of ama1 and mdr1 to track within-host P. falciparum diversity throughout treatment in a clinical drug trial
Kevin Wamae, Leonard Ndwiga, Oksana Kharabora, Kelvin Kimenyi, Victor Osoti, Zaydah de Laurent, Juliana Wambua, Jennifer Musyoki, Caroline Ngetsa, Peter Kalume, Gabriel Mwambingu, Mainga Hamaluba, Rob van der Pluijm, Arjen M Dondorp, Jeffrey Bailey, Jonathan Juliano, Philip Bejon, Lynette Ochola-Oyier
Wellcome Open Research 2024;7;95
DOI: 10.12688/wellcomeopenres.17736.2
scRNA-Seq reveals elevated interferon responses and TNF-α signaling via NFkB in monocytes in children with uncomplicated malaria
Collins M. Morang’a, Riley S. Drake, Vincent N. Miao, Nancy K. Nyakoe, Dominic S.Y. Amuzu, Vincent Appiah, Yaw Aniweh, Yaw Bediako, Saikou Y. Bah, Alex K. Shalek, Gordon A. Awandare, Thomas D. Otto, Lucas Amenga–Etego
medRxiv 2024
DOI: 10.1101/2023.06.02.23290878
Regional Plasmodium falciparum subpopulations and malaria transmission connectivity in Africa detected with an enlarged panel of genome-wide microsatellite loci.
Martha Anita Demba, Edwin Kamau, Jaishree Raman, Karim Mane, Lucas Emenga-Etego, Tobias Apinjo, Deus Isheghoma, Lemu Golassa, Oumou Maiga, Anita Ghansah, Marielle Bouyou-Akotet, William Yavo, Milijoana Randrianarivelojosia, Fadel Muhammadou Diop, Eniyou Oriero, David Jeffries, Umberto D’Alessandro, Abdoulaye Djimde, Alfred Amambua-Ngwa
bioRxiv 2024
DOI: 10.1101/2024.03.08.584049
Role for gene conversion in the evolution of cell-surface antigens of the malaria parasite Plasmodium falciparum
Brice Letcher, Sorina Maciuca, Zamin Iqbal
PLOS Biology 2024;22(3);e3002507
DOI: 10.1371/journal.pbio.3002507
Evaluating evidence for co-geography in the Anopheles–Plasmodium host-parasite system
Clara T Rehmann, Peter L Ralph, Andrew D Kern
G3 Genes Genomes Genetics 2024; 14;(3)
DOI: 10.1093/g3journal/jkae008
Plasmodium falciparum transmission in the highlands of Ethiopia is driven by closely related and clonal parasites
Aurel Holzschuh, Yalemwork Ewnetu, Lise Carlier, Anita Lerch, Inna Gerlovina, Sarah Cate Baker, Delenasaw Yewhalaw, Werissaw Haileselassie, Nega Berhane, Wossenseged Lemma, Cristian Koepfli
Mol Ecol 2024;33(6);e17292
DOI: 10.1111/mec.17292
Artemisinin resistance-associated gene mutations in Plasmodium falciparum: A case study of severe malaria from Mozambique
Daniela Casanova, Vitória Baptista, Magda Costa, Bruno Freitas, Maria das Neves Imaculada Pereira, Carla Calçada, Paula Mota, Olena Kythrich, Maria Helena Jacinto Sarmento Pereira, Nuno S. Osório, Maria Isabel Veiga
Travel Medicine and Infectious Disease 2024;57;102684
DOI: 10.1016/j.tmaid.2023.102684
A novel computational pipeline for var gene expression augments the discovery of changes in the Plasmodium falciparum transcriptome during transition from in vivo to short-term in vitro culture
Clare Andradi-Brown, Jan Stephan Wichers-Misterek, Heidrun von Thien, Yannick D Höppner, Judith AM Scholz, Helle Hansson, Emma Filtenborg Hocke, Tim Wolf Gilberger, Michael F Duffy, Thomas Lavstsen, Jake Baum, Thomas D Otto, Aubrey J Cunnington ,Anna Bachmann
eLife 2024;25;12:RP87726
DOI: 10.7554/eLife.87726.3
The Kelch13 compartment contains highly divergent vesicle trafficking proteins in malaria parasites
Sabine Schmidt, Jan Stephan Wichers-Misterek, Hannah Michaela Behrens, Jakob Birnbaum, Isabelle G. Henshall, Jana Dröge, Ernst Jonscher, Sven Flemming, Carolina Castro-Peña, Paolo Mesén-Ramírez, Tobias Spielmann
PLoS Pathog 2023;19(12);e1011814
DOI: 10.1371/journal.ppat.1011814
Genomic analysis of Plasmodium vivax describes patterns of connectivity and putative drivers of adaptation in Ethiopia
Alebachew Messele Kebede , Edwin Sutanto , HidayatTrimarsanto, Ernest Diez Benavente , Mariana Barnes, Richard D. Pearson, Sasha V. Siegel , Berhanu Erko, Ashenafi Assefa, Sisay Getachew, Abraham Aseffa , Beyene Petros, Eugenia Lo, Rezika Mohammed, Daniel Yilma, Angela Rumaseb , Francois Nosten, Rintis Noviyanti , Julian C. Rayner, Dominic P. Kwiatkowski, Ric N. Price, LemuGolassa & Sarah Auburn
Scientific Reports 2023;13;20788
DOI: 10.1038/s41598-023-47889-w
Genomics of Plasmodium vivax in Colombia reveals evidence of local bottle-necking and inter-country connectivity in the Americas
Edwin Sutanto, Zuleima Pava, Diego F. Echeverry, Tatiana M. Lopera‑Mesa, Lidia Madeline Montenegro, Maria F.Yasnot‑Acosta, Ernest Diez Benavente, Richard D. Pearson, Sócrates Herrera, Myriam Arévalo‑Herrera, Hidayat Trimarsanto, Angela Rumaseb , Rintis Noviyanti, Dominic P. Kwiatkowski, Ric N. Price & Sarah Auburn
Scientific Reports 2023;13;19779
DOI: 10.1038/s41598-023-46076-1
Mapping the genomic landscape of multidrug resistance in Plasmodium falciparum and its impact on parasite fitness.
Sachel Mok, Tomas Yeo, Davin Hong, Melanie J. Shears, Leila S. Ross, Kurt E. Ward, Satish K. Dhingra, Mariko Kanai, Jessica L. Bridgford, Abhai K. Tripathi, Godfree Mlambo, Anna Y. Burkhard, Megan R. Ansbro, Kate J. Fairhurst, Eva Gil-Iturbe, Heekuk Park, Felix D. Rozenberg, Jonathan Kim, Filippo Mancia, Rick M. Fairhurst, Matthias Quick, Anne-Catrin Uhlemann, Photini Sinnis, David A. Fidock
Science Advances 2023;9(45);eadi2364
DOI: 10.1126/sciadv.adi2364
Evolution and spread of Plasmodium falciparum mutations associated with resistance to sulfadoxine-pyrimethamine in central Africa: a cross-sectional study
Emilie Guémas, Romain Coppée, Sandie Ménard, Milena du Manoir, Sandrine Nsango, Dieudonné Makaba Mvumbi, Emmanuel Nakoune, Carole Else Eboumbou Moukoko, Marielle Karine Bouyou Akotet, Tatfeng Youtchou Mirabeau, Sylvie Manguin, Doudou Malekita Yobi, Jean Akiana, Lady Charlène Kouna, Denise Patricia Mawili Mboumba, Dominique Fatima Voumbo-Matoumona, Alliance-Laure Otam, Pierre-Alain Rubbo, Jean-Pierre Lombart, Elisabeth Kwanai, Olivia Cohen, Xavier Iriart, Lawrence Ayong, Jean Bernard Lekana-Douki, Frédéric Ariey, Antoine Berry
Lancet Microbe 2023;4(12);e983-e993
DOI: 10.1016/ S2666-5247(23)00211-2
MalariaSED: a deep learning framework to decipher the regulatory contributions of noncoding variants in malaria parasites
Chengqi Wang, Yibo Dong, Chang Li, Jenna Oberstaller, Min Zhang, Justin Gibbons, Camilla Valente Pires, Mianli Xiao, Lei Zhu, Rays H. Y. Jiang, Kami Kim, Jun Miao, Thomas D. Otto, Liwang Cui, John H. Adams & Xiaoming Liu
Genome Biology 2023;24(231)
DOI: 10.1186/s13059-023-03063-z
Molecular surveillance of Plasmodium falciparum drug-resistance markers in Vietnam using multiplex amplicon sequencing (2000–2016)
Eduard Rovira-Vallbona, Johanna Helena Kattenberg, Nguyen Van Hong, Pieter Guetens, Hideo Imamura, Pieter Monsieurs, Driss Chiheb, Annette Erhart, Bui Quang Phuc, Nguyen Xuan Xa & Anna Rosanas-Urgell
Scientific Reports 2023;13;13948
DOI: 10.1038/s41598-023-40935-7
A potent and durable malaria transmission-blocking vaccine designed from a single-component 60-copy Pfs230D1 nanoparticle
Nichole D. Salinas, Rui Ma, Thayne H. Dickey, Holly McAleese, Tarik Ouahes, Carole A. Long, Kazutoyo Miura, Lynn E. Lambert & Niraj H. Tolia
npj Vaccines 2023;8(124)
DOI: 10.1038/s41541-023-00709-8
Genomic analysis of Plasmodium falciparum isolates across different altitudinal zones along the slope of Mount Cameroon
Tobias O. Apinjoh, Marcelus U. Ajonina, Deriba Abera, Hanesh F. Chi, Roland B. Tata, Regina N. Mugri, Lemu Golassa, Eric A. Achidi, Alfred Amambua-Ngwa
Frontiers in Malaria 2023;1
DOI: 10.3389/fmala.2023.1075755
An optimized GATK4 pipeline for Plasmodium falciparum whole genome sequencing variant calling and analysis
Karamoko Niaré, Bryan Greenhouse & Jeffrey A. Bailey
Malaria Journal 2023;22(207)
DOI: 10.1186/s12936-023-04632-0
Highly multiplexed ddPCR-amplicon sequencing reveals strong Plasmodium falciparum population structure and isolated populations amenable to local elimination efforts in Zanzibar
Aurel Holzschuh, Anita Lerch, Inna Gerlovina, Bakar S. Fakih, Abdul-wahid H. Al-mafazy, Erik J. Reaves, Abdullah Ali, Faiza Abbas, Mohamed Haji Ali, Mohamed Ali Ali, Manuel W. Hetzel, Joshua Yukich & Cristian Koepfli
Nat Commun 2023;14(3699)
DOI: 10.1038/s41467-023-39417-1
Genome-wide genetic variation and molecular surveillance of drug resistance in Plasmodium falciparum isolates from asymptomatic individuals in Ouélessébougou, Mali
Leen N Vanheer, Almahamoudou Mahamar, Emilia Manko, Sidi M Niambele, Koualy Sanogo, Ahamadou Youssouf, Adama Dembele, Makonon Diallo, Seydina O Maguiraga, Jody Phelan, Ashley Osborne, Anton Spadar, Merel J Smit, Teun Bousema, Chris Drakeley, Taane G Clark, William Stone, Alassane Dicko, Susana Campino
Scientific Reports 2023;13(1);9522
DOI: 10.1038/s41598-023-36002-w.
coiaf: Directly estimating complexity of infection with allele frequencies
Aris Paschalidis, Oliver J. Watson, Ozkan Aydemir, Robert Verity , Jeffrey A. Bailey
PLoS Comput Biol 2023;19(6);e101024
DOI: 10.1371/journal.pcbi.1010247
Targeted and whole-genome sequencing reveal a north-south divide in P. falciparum drug resistance markers and genetic structure in Mozambique
Clemente da Silva, Simone Boene, Debayan Datta, Eduard Rovira-Vallbona, Andrés Aranda-Díaz, Pau Cisteró, Nicholas Hathaway, Sofonias Tessema, Arlindo Chidimatembue, Glória Matambisso, Abel Nhama, Eusebio Macete, Arnau Pujol, Lidia Nhamussua, Beatriz Galatas, Caterina Guinovart, Sónia Enosse, Eva De Carvalho, Eric Rogier, Mateusz M. Plucinski, James Colborn, Rose Zulliger, Abuchahama Saifodine, Pedro L. Alonso, Baltazar Candrinho, Bryan Greenhouse, Pedro Aide, Francisco Saute & Alfredo Mayor
Nature Communications Biology 2023;6;619
DOI: 10.1038/s42003-023-04997-7
Performance of SNP barcodes to determine genetic diversity and population structure of Plasmodium falciparum in Africa
Dionne C Argyropoulos, Mun Hua Tan, Courage Adobor, Benedicta Mensah, Frédéric Labbé, Kathryn E Tiedje, Kwadwo A Koram, Anita Ghansah, Karen P Day
Frontiers in Genetics 2023;13
DOI: 10.3389/fgene.2023.1071896
Genomic analysis of Indian isolates of Plasmodium falciparum: Implications for drug resistance and virulence factors
Deepak Choubey, Bhagyashree Deshmukh, Anjani Gopal Rao, Abhishek Kanyal, Amiya Kumar Hati, Somenath Roy, Krishanpal Karmodiya
Int J Parasitol Drugs Drug Resist 2023;22;52–60
DOI: 10.1016/j.ijpddr.2023.05.003
MinSNPs: an R package for derivation of resolution-optimised SNP sets from microbial genomic data
Hoon KS, Holt DC, Auburn S, Shaw P, Giffard PM
PeerJ 2023;11;e15339
DOI: 10.7717/peerj.15339
Genomic variation during culture adaptation of genetically complex Plasmodium falciparum clinical isolates
Claessens A, Stewart LB, Drury E, Ahouidi AD, Amambua-Ngwa A, Diakite M, Kwiatkowski DP, Awandare GA, Conway DJ.
Microbial Genomics 2023;9(5)
DOI: 10.1099/mgen.0.001009
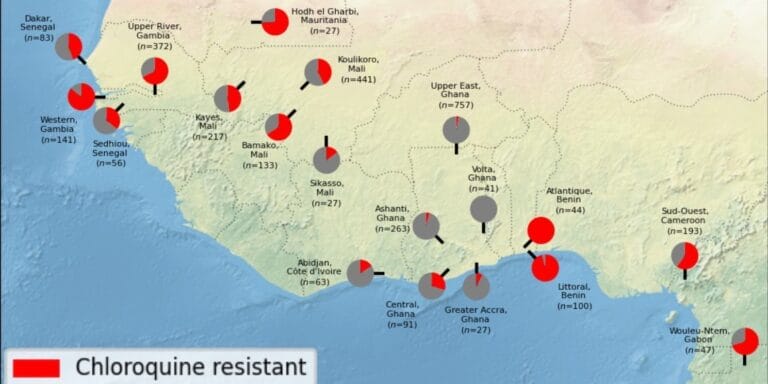
Chloroquine resistance evolution in Plasmodium falciparum is mediated by the putative amino acid transporter AAT1
Alfred Amambua-Ngwa, Katrina A. Button-Simons, Xue Li , Sudhir Kumar , et al
Nature Microbiology 2023;8;1213–1226
DOI: https://doi.org/10.1038/s41564-023-01377-z
Population dynamics and drug resistance mutations in Plasmodium falciparum on the Bijagós Archipelago, Guinea-Bissau
Sophie Moss, Emilia Mańko, Hristina Vasileva, Eunice Teixeira Da Silva, Adriana Goncalves, Ashley Osborne, Jody Phelan, Amabelia Rodrigues, Paulo Djata, Umberto D'Alessandro, David Mabey, Sanjeev Krishna, Anna Last, Taane G Clark, Susana Campino
Scientific Reports 2023;13(1);6311
DOI: 10.1038/s41598-023-33176-1
Characterization of a novel Plasmodium falciparum merozoite surface antigen and potential vaccine target
Karamoko Niaré, Timothy Chege, Micha Rosenkranz, Kennedy Mwai, Zoe Saßmannshausen, Dennis Odera, Lydia Nyamako, James Tuju, Tiono Alfred, John N Waitumbi, Bernhards Ogutu, Sodiomon B Sirima, Gordon Awandare, Bourema Kouriba, Julian C Rayner, Faith H A Osier
Frontiers Immunology 2023;14;1156806
DOI: 10.3389/fimmu.2023.1156806
Population Genomic Evidence of Adaptive Response during the Invasion History of Plasmodium falciparum in the Americas
Margaux J M Lefebvre, Josquin Daron, Eric Legrand, Michael C Fontaine, Virginie Rougeron, Franck Prugnolle
Mol Biol Evol 2023;40(5);msad082
DOI: 10.1093/molbev/msad082
Potent acyl-CoA synthetase 10 inhibitors kill Plasmodium falciparum by disrupting triglyceride formation
Selina Bopp, Charisse Flerida A. Pasaje, Robert L. Summers, Pamela Magistrado-Coxen, Kyra A. Schindler, Victoriano Corpas-Lopez, Tomas Yeo, Sachel Mok, Sumanta Dey, Sebastian Smick, Armiyaw S. Nasamu, Allison R. Demas, Rachel Milne, Natalie Wiedemar, Victoria Corey, Maria De Gracia Gomez-Lorenzo, Virginia Franco, Angela M. Early, Amanda K. Lukens, Danny Milner, Jeremy Furtado, Francisco-Javier Gamo, Elizabeth A. Winzeler, Sarah K. Volkman, Maëlle Duffey, Benoît Laleu, David A. Fidock, Susan Wyllie, Jacquin C. Niles, Dyann F. Wirth
Nat Commun 2023;14(1455)
DOI: 10.1038/s41467-023-36921-2
Genetic diversity and natural selection of rif gene (PF3D7_1254800) in the Plasmodium falciparum global populations
Shao-Jie Xu, Hai-Mo Shen, Yan-Bing Cui, Shen-Bo Chen, Bin Xu, Jun-Hu Chen
Mol Biochem Parasitol 2023;254;111558
DOI: 10.1016/j.molbiopara.2023.111558
Limited genetic variations of the Rh5-CyRPA-Ripr invasion complex in Plasmodium falciparum parasite population in selected malaria-endemic regions, Kenya
Harrison Waweru, Bernard N. Kanoi, Josiah O. Kuja, Mary Maranga, James Kongere, Michael Maina, Johnson Kinyua and Jesse Gitaka
Frontiers in Tropical Disease 2023;4;1102265
DOI: 10.3389/fitd.2023.1102265
Malaria Molecular Surveillance in the Peruvian Amazon with a Novel Highly Multiplexed Plasmodium falciparum AmpliSeq Assay
Johanna Helena Kattenberg, Carlos Fernandez-Miñope, Norbert J. van Dijk, Lidia Llacsahuanga Allcca, Pieter Guetens, Hugo O. Valdivia, Jean-Pierre Van geertruyden, Eduard Rovira-Vallbona, Pieter Monsieurs, Christopher Delgado-Ratto, Dionicia Gamboa, Anna Rosanas-Urgell
Microbiology Spectrum 2023;11;e00960-22
DOI: 10.1128/spectrum.00960-22
Nosocomial Malaria Transmissions Resolved by Genomic Analyses—A Retrospective Case Report Study in France: 2007–2021
Romain Coppée, Véronique Sarrasin, Rizwana Zaffaroulah, Azza Bouzayene, Marc Thellier, Harold Noël, Jérôme Clain, Sandrine Houzé, the Investigation Study Group
Clinical Infectious Diseases 2023;76(4);631–639
DOI: 10.1093/cid/ciac813
Potent transmission-blocking monoclonal antibodies from naturally exposed individuals target a conserved epitope on Plasmodium falciparum Pfs230
Danton Ivanochko, Amanda Fabra-García, Karina Teelen, Marga van de Vegte-Bolmer, Geert-Jan van Gemert, Jocelyn Newton, Anthony Semesi, Marloes de Bruijni, Judith Bolscher, Jordache Ramjith, Marta Szabat, Stefanie Vogt, Lucas Kraft, Sherie Duncan, Shwu-Maan Lee, Moses R. Kamya, Margaret E. Feeney, Prasanna Jagannathan, Bryan Greenhouse, Robert W. Sauerwein, C. Richter King, Randall S. MacGill, Teun Bousema, Matthijs M. Jore, Jean-Philippe Julien
Immunity 2023;56(2);420–432
DOI: 10.1016/j.immuni.2023.01.013
Short tandem repeat polymorphism in the promoter region of cyclophilin 19B drives its transcriptional upregulation and contributes to drug resistance in the malaria parasite Plasmodium falciparum
Kucharski M, Wirjanata G, Nayak S, Boentoro J, Dziekan JM, Assisi C, van der Pluijm RW, Miotto O, Mok S, Dondorp AM, Bozdech Z.
PLOS Pathogens 2023;19(1);e1011118
DOI: 10.1371/journal.ppat.1011118
Role of Pfs47 in the dispersal of ancestral Plasmodium falciparum malaria through adaptation to different anopheline vectors
Molina-Cruz A, Canepa GE, Dwivedi A, Liu W, Raytselis N, Antonio-Nkondjio C, Hahn BH, Silva JC, Barillas-Mury C.
PNAS 2023;120(5);e2213626120
DOI: 10.1073/pnas.2213626120
Genetic Epidemiology Use Cases for Malaria Control Programmes: A Methodology
Olivo Miotto, Principal Investigator, GenRe-Mekong Project
Genetic Epidemiology Use Cases for Malaria Control Programmes: A Methodology 2023
DOI:
Unravelling var complexity: Relationship between DBLα types and var genes in Plasmodium falciparum
Mun Hua Tan, Heejung Shim, Yao-ban Chan, Karen P. Day
Front Parasitol 2023;1;1006341
DOI: 10.3389/fpara.2022.1006341
Drug Resistant Prediction Based on Plasmodium falciparum DNA-Barcoding using Bidirectional Long Short Term Memory Method
Lailil Muflikhah, Nashi Widodo, Novanto Yudistira, Achmad Ridok
International Journal of Advanced Computer Science and Applications (IJACSA) 2023;14(7)
DOI: 10.14569/IJACSA.2023.0140747
Drug resistance and population structure of Plasmodium falciparum and Plasmodium vivax in the Peruvian Amazon
Villena FE, Sanchez JF, Nolasco O, Braga G, Ricopa L, Barazorda K, Salas CJ, Lucas C, Lizewski SE, Joya CA, Gamboa D, Delgado-Ratto C, Valdivia HO
Scientific Reports 2022;12(1);16474
DOI: 10.1038/s41598-022-21028-3
Phenotypic characterization of Ghanaian P. falciparum clinical isolates reveals a homogenous parasite population
Laty G. Thiam, Prince B. Nyarko, Felix Ansah, Makhtar Niang, Gordon A. Awandare, Yaw Aniweh
Frontiers Immunology 2022;13;1009252
DOI: 10.3389/fimmu.2022.1009252
Design and implementation of multiplexed amplicon sequencing panels to serve genomic epidemiology of infectious disease: A malaria case study
LaVerriere E, Schwabl P, Carrasquilla M, Taylor AR, Johnson ZM, Shieh M, Panchal R, Straub TJ, Kuzma R, Watson S, Buckee CO, Andrade CM, Portugal S, Crompton PD, Traore B, Rayner JC, Corredor V, James K, Cox H, Early AM, MacInnis BL, Neafsey DE.
Molecular Ecology Resources 2022;22;2285
DOI: https://doi.org/10.1111/1755-0998.13622
Molecular epidemiology of resistance to antimalarial drugs in the Greater Mekong subregion: an observational study
Imwong M, Dhorda M, Myo Tun K, Thu AM, Phyo AP, Proux S, Suwannasin K, Kunasol C, Srisutham S, Duanguppama J, Vongpromek R, Promnarate C, Saejeng A, Khantikul N, Sugaram R, Thanapongpichat S, Sawangjaroen N, Sutawong K, Han KT, Htut Y, Linn K, Win AA, Hlaing TM, van der Pluijm RW, Mayxay M, Pongvongsa T, Phommasone K, Tripura R, Peto TJ, von Seidlein L, Nguon C, Lek D, Chan XHS, Rekol H, Leang R, Huch C, Kwiatkowski DP, Miotto O, Ashley EA, Kyaw MP, Pukrittayakamee S, Day NPJ, Dondorp AM, Smithuis FM, Nosten FH, White NJ.
The Lancet Infectious Diseases 2020;20(12);1470-1480
DOI: https://doi.org/10.1016/S1473-3099(20)30228-0
Protective Immunity against Severe Malaria in Children Is Associated with a Limited Repertoire of Antibodies to Conserved PfEMP1 Variants
Tessema SK, Nakajima R, Jasinskas A, Monk SL, Lekieffre L, Lin E, Kiniboro B, Proietti C, Siba P, Felgner PL, Doolan DL, Mueller I, Barry AE.
Cell Host & Microbe 2019;26(5);579-590.e5
DOI: https://doi.org/10.1016/j.chom.2019.10.012
An open dataset of Plasmodium vivax genome variation in 1,895 worldwide samples
MalariaGEN, Ishag Adam, Mohammad Shafiul Alam, Sisay Alemu, Chanaki Amaratunga, Roberto Amato, Voahangy Andrianaranjaka, Nicholas M Anstey, Abraham Aseffa, Elizabeth Ashley, Ashenafi Assefa, Sarah Auburn, Bridget E Barber, Alyssa Barry, Dhelio Batista Pereira, Jun Cao, Nguyen Hoang Chau, Kesinee Chotivanich, Cindy Chu, Arjen M. Dondorp, Eleanor Drury, Diego F. Echeverry, Berhanu Erko, Fe Espino, Rick M. Fairhurst, Abdul Faiz, María Fernanda Villegas, Qi Gao, Lemu Golassa, Sonia Goncalves, Matthew J Grigg, Yaghoob Hamedi, Tran Tinh Hien, Ye Htut, Kimberly J Johnson, Nadira Karunaweera, Wasif Khan, Srivicha Krudsood, Dominic P Kwiatkowski, Marcus Lacerda, Benedikt Ley, Pharath Lim, Yaobao Liu, Alejandro Llanos-Cuentas, Chanthap Lon, Tatiana Lopera-Mesa, Jutta Marfurt, Pascal Michon, Olivo Miotto, Rezika Mohammed, Ivo Mueller, Chayadol Namaik-larp, Paul N Newton, Thuy-Nhien Nguyen, François Nosten, Rintis Noviyanti , Zuleima Pava, Richard D Pearson, Beyene Petros, Aung P Phyo, Ric N Price, Sasithon Pukrittayakamee , Awab Ghulam Rahim, Milijaona Randrianarivelojosia, Julian C Rayner Orcid, Angela Rumaseb, Sasha V Siegel, Victoria J Simpson, Kamala Thriemer, Alberto Tobon-Castano, Hidayat Trimarsanto, Marcelo Urbano Ferreira, Ivan D Vélez, Sonam Wangchuk, Thomas E Wellems, Nicholas J White, Timothy William, Maria F Yasnot, Daniel Yilma
Wellcome Open Res 2022;7:136
DOI: 10.12688/wellcomeopenres.17795.1
Artemisinin resistance in the malaria parasite, Plasmodium falciparum, originates from its initial transcriptional response
Zhu L, van der Pluijm RW, Kucharski M, Nayak S, Tripathi J, White NJ, Day NPJ, Faiz A, Phyo AP, Amaratunga C, Lek D, Ashley EA, Nosten F, Smithuis F, Ginsburg H, von Seidlein L, Lin K, Imwong M, Chotivanich K, Mayxay M, Dhorda M, Nguyen HC, Nguyen TNT, Miotto O, Newton PN, Jittamala P, Tripura R, Pukrittayakamee S, Peto TJ, Hien TT, Dondorp AM, Bozdech Z.
Communications Biology 2022;5;274
DOI: https://doi.org/10.1038/s42003-022-03215-0
Resolving drug selection and migration in an inbred South American Plasmodium falciparum population with identity-by-descent analysis
Carrasquilla M, Early AM, Taylor AR, Knudson A, Echeverry DF, Anderson TJ, Mancilla E, Aponte S, Cárdenas P, Buckee CO, Rayner JC, Sáenz FE, Neafsey DE, Corredor V
bioRxiv 2022
DOI: https://doi.org/10.1101/2022.02.18.480973
Global diversity and balancing selection of 23 leading Plasmodium falciparum candidate vaccine antigens
Myo T. Naung, Elijah Martin, Jacob Munro, Somya Mehra, Andrew J. Guy, Moses Laman, G. L. Abby Harrison, Livingstone Tavul, Manuel Hetzel, Dominic Kwiatkowski, Ivo Mueller, Melanie Bahlo, Alyssa E. Barry
PLoS Comput Biol 2022;18:2;e1009801
DOI: 10.1371/journal.pcbi.1009801
Increase in the proportion of Plasmodium falciparum with kelch13 C580Y mutation and decline in pfcrt and pfmdr1 mutant alleles in Papua New Guinea
Yoshida N, Yamauchi M, Morikawa R, Hombhanje F, Mita T.
Malaria Journal 2021;20;410
DOI: https://doi.org/10.1186/s12936-021-03933-6
Genetic surveillance in the Greater Mekong Subregion and South Asia to support malaria control and elimination: about the data
Jacob CG, Thuy-Nhien N, Mayxay M, et al.
eLife 2021;10:e62997
DOI: 10.7554/eLife.62997
Using deep learning to identify recent positive selection in malaria parasite sequence data
Deelder W, Benavente ED, Phelan J, Manko E, Campino S, Palla L, Clark TG.
Malaria Journal 2021;20(1);270
DOI: 10.1186/s12936-021-03788-x
Arterolane-piperaquine-mefloquine versus arterolane-piperaquine and artemether-lumefantrine in the treatment of uncomplicated Plasmodium falciparum malaria in Kenyan children: a single-centre, open-label, randomised, non-inferiority trial
Mainga Hamaluba, Rob W van der Pluijm, Joseph Weya, Patricia Njuguna, Mwanajuma Ngama, Peter Kalume, Gabriel Mwambingu, Caroline Ngetsa, Juliana Wambua, Mwanamvua Boga, Neema Mturi, Altaf A Lal, Arshad Khuroo, Walter R J Taylor, Sónia Gonçalves, Olivo Miotto, Mehul Dhorda, Brian Mutinda, Mavuto Mukaka, Naomi Waithira, Richard M Hoglund, Mallika Imwong, Joel Tarning, Nicholas P J Day, Nicholas J White, Philip Bejon, Arjen M Dondorp
The Lancet Infectious Diseases 2021;21(10);1395-1406
DOI: 10.1016/S1473-3099(20)30929-4
Distinctive genetic structure and selection patterns in Plasmodium vivax from South Asia and East Africa
Benavente ED, Manko E, Phelan J, Campos M, Nolder D, Fernandez D, Velez-Tobon G, Castaño AT, Dombrowski JG, Marinho CRF, Aguiar ACC, Pereira DB, Sriprawat K, Nosten F, Moon R, Sutherland CJ, Campino S, Clark TG.
Nature Communications 2021;12(1);3160
DOI: 10.1038/s41467-021-23422-3
Population genomic evidence of Plasmodium vivax Southeast Asian origin
Daron J, Boissière A, Boundenga L, Ngoubangoye B, Houze S, Arnathau C, Sidobre C, Trape JF, Durand P, Renaud F, Fontaine MC, Prugnolle F, Rougeron V.
Science Advances 2021;7(18);eabc3713
DOI: 10.1126/sciadv.abc3713
Genomic analysis reveals independent evolution of Plasmodium falciparum populations in Ethiopia
Abera D, Kibet CK, Degefa T, Amenga-Etego L, Bargul JL, Golassa L.
Malaria Journal 2021;20;129
DOI: https://doi.org/10.1186/s12936-021-03660-y
Temporal evolution of sulfadoxine-pyrimethamine resistance genotypes and genetic diversity in response to a decade of increased interventions against Plasmodium falciparum in northern Ghana
Lucas N. Amenga-Etego, Victor Asoala, Godfred Agongo, Christopher Jacob, Sonia Goncalves, Gordon A. Awandare, Kirk A. Rockett & Dominic Kwiatkowski
Malaria Journal 2021;20(152)
DOI: 10.1186/s12936-021-03693-3
An open dataset of Plasmodium falciparum genome variation in 7,000 worldwide samples
MalariaGEN, Ambroise Ahouidi, Mozam Ali, Jacob Almagro-Garcia, Alfred Amambua-Ngwa, Chanaki Amaratunga, Roberto Amato,Lucas Amenga-Etego, Ben Andagalu, Tim J. C. Anderson, Voahangy Andrianaranjaka, Tobias Apinjoh, Cristina Ariani, Elizabeth A. Ashley, Sarah Auburn, Gordon Awandare, Hampate Ba, Vito Baraka, Alyssa E. Barry1, Philip Bejon, Gwladys I. Bertin, Maciej F. Boni, Steffen Borrmann, Teun Bousema, Oralee Branch, Peter C. Bull, George B. J. Busby, Thanat Chookajorn, Kesinee Chotivanich, Antoine Claessens, David Conway, Alister Craig, Umberto D'Alessandro, Souleymane Dama, Nicholas Day, Brigitte Denis, Mahamadou Diakite, Abdoulaye Djimdé, Christiane Dolecek, Arjen Dondorp, Chris Drakeley, Eleanor Drury, Patrick Duffy, Diego F. Echeverry, Thomas G. Egwang, Berhanu Erko, Rick M. Fairhurst, Abdul Faiz, Caterina A. Fanello, Mark M. Fukuda, Dionicia Gamboa, Anita Ghansah, Lemu Golassa, Sonia Goncalves, William L. Hamilton, G. L. Abby Harrison, Lee Hart, Christa Henrichs, Tran Tinh Hien, Catherine A. Hill, Abraham Hodgson, Christina Hubbart, Mallika Imwong, Deus S. Ishengoma, Scott A. Jackson, Chris G. Jacob, Ben Jeffery, Anna E. Jeffreys, Kimberly J. Johnson, Dushyanth Jyothi, Claire Kamaliddin, Edwin Kamau, Mihir Kekre, Krzysztof Kluczynski, Theerarat Kochakarn, Abibatou Konaté, Dominic P. Kwiatkowski, Myat Phone Kyaw, Pharath Lim, Chanthap Lon, Kovana M. Loua, Oumou Maïga-Ascofaré, Cinzia Malangone, Magnus Manske, Jutta Marfurt, Kevin Marsh, Mayfong Mayxay, Alistair Miles, Olivo Miotto, Victor Mobegi, Olugbenga A. Mokuolu, Jacqui Montgomery, Ivo Mueller, Paul N. Newton, Thuy Nguyen, Thuy-Nhien Nguyen, Harald Noedl, Francois Nosten, Rintis Noviyanti, Alexis Nzila, Lynette I. Ochola-Oyier, Harold Ocholla, Abraham Oduro, Irene Omedo, Marie A. Onyamboko, Jean-Bosco Ouedraogo, Kolapo Oyebola, Richard D. Pearson, Norbert Peshu, Aung Pyae Phyo, Chris V. Plowe, Ric N. Price, Sasithon Pukrittayakamee, Milijaona Randrianarivelojosia, Julian C. Rayner, Pascal Ringwald, Kirk A. Rockett, Katherine Rowlands, Lastenia Ruiz, David Saunders, Alex Shayo, Peter Siba, Victoria J. Simpson, Jim Stalker, Xin-zhuan Su, Colin Sutherland, Shannon Takala-Harrison, Livingstone Tavul, Vandana Thathy, Antoinette Tshefu, Federica Verra, Joseph Vinetz, Thomas E. Wellems, Jason Wendler, Nicholas J. White, Ian Wright, William Yavo, Htut Ye.
Wellcome Open Research 2021;6:42
DOI: https://doi.org/10.12688/wellcomeopenres.16168.1
Emergence of artemisinin-resistant Plasmodium falciparum with kelch13 C580Y mutations on the island of New Guinea
Miotto O, Sekihara M, Tachibana SI, Yamauchi M, Pearson RD, Amato R, Gonçalves S, Mehra S, Noviyanti R, Marfurt J, Auburn S, Price RN, Mueller I, Ikeda M, Mori T, Hirai M, Tavul L, Hetzel MW, Laman M, Barry AE, Ringwald P, Ohashi J, Hombhanje F, Kwiatkowski DP, Mita T.
PLoS Pathogens 2020;16(12);e1009133
DOI: https://doi.org/10.1371/journal.ppat.1009133
Genomic surveillance of Plasmodium falciparum and Plasmodium vivax cases at the University Hospital in Tegucigalpa, Honduras
Hugo O Valdivia, Fredy E Villena, Stephen E Lizewski, Jorge Garcia, Jackeline Alger, Danett K Bishop
Scientific Reports 2020;10 (1);20975
DOI: 10.1038/s41598-020-78103-w
Identity-by-descent with uncertainty characterises connectivity of Plasmodium falciparum populations on the Colombian-Pacific coast
Taylor AR, Echeverry DF, Anderson TJC, Neafsey DE, Buckee CO.
PLoS Genetics 2020;16(11);e1009101
DOI: 10.1371/journal.pgen.1009101
Whole genome sequencing of Plasmodium vivax isolates reveals frequent sequence and structural polymorphisms in erythrocyte binding genes
Ford A, Kepple D, Abagero BR, Connors J, Pearson R, Auburn S, Getachew S, Ford C, Gunalan K, Miller LH, Janies DA, Rayner JC, Yan G, Yewhalaw D, Lo E.
PLoS Neglected Tropical Diseases 2020;14(10);e0008234
DOI: 10.1371/journal.pntd.0008234
Antimalarial drug resistance molecular makers of Plasmodium falciparum isolates from Sudan during 2015–2017
Hussien M, Abdel Hamid MM, Elamin EA, Hassan AO, Elaagip AH, Salama AHA, Abdelraheem MH, Mohamed AO.
PLOS ONE 2020;15(8);e0235401
DOI: 10.1371/journal.pone.0235401
Occurrence of septuple and elevated Pfdhfr-Pfdhps quintuple mutations in a general population threatens the use of sulfadoxine-pyrimethamine for malaria prevention during pregnancy in eastern coast of Tanzania
George M. Bwire, Wigilya P. Mikomangwa and Manase Kilonz
BMC Infectious Diseases 2020;20;530
DOI: 10.1186/s12879-020-05253-7
Genetic diversity and neutral selection in Plasmodium vivax erythrocyte binding protein correlates with patient antigenicity
Han JH, Cho JS, Ong JJY, Park JH, Nyunt MH, Sutanto E, Trimarsanto H, Petros B, Aseffa A, Getachew S, Sriprawat K, Anstey NM, Grigg MJ, Barber BE, William T, Qi G, Liu Y, Pearson RD, Auburn S, Price RN, Nosten F, Rénia L, Russell B, Han ET.
PLoS Neglected Tropical Diseases 2020;14(7);e0008202
DOI: 10.1371/journal.pntd.0008202
Efficacy of dihydroartemisinin/piperaquine and artesunate monotherapy for the treatment of uncomplicated Plasmodium falciparum malaria in Central Vietnam
Rovira-Vallbona E, Van Hong N, Kattenberg JH, Huan RM, Hien NTT, Ngoc NTH, Guetens P, Hieu NL, Mai TT, Duong NTT, Duong TT, Phuc BQ, Xa NX, Erhart A, Rosanas-Urgell A.
Journal of Antimicrobial Chemotherapy 2020;75;2272
DOI: https://doi.org/10.1093/jac/dkaa172
Mapping the travel patterns of people with malaria in Bangladesh
Sinha I, Sayeed AA, Uddin D, Wesolowski A, Zaman SI, Faiz MA, Ghose A, Rahman MR, Islam A, Karim MJ, Saha A, Rezwan MK, Shamsuzzaman AKM, Jhora ST, Aktaruzzaman MM, Chang HH, Miotto O, Kwiatkowski D, Dondorp AM, Day NPJ, Hossain MA, Buckee C, Maude RJ.
BMC Medicine 2020;18(1);45
DOI: 10.1186/s12916-020-1512-5
Plasmodium vivax Malaria Viewed through the Lens of an Eradicated European Strain
van Dorp L, Gelabert P, Rieux A, de Manuel M, de-Dios T, Gopalakrishnan S, Carøe C, Sandoval-Velasco M, Fregel R, Olalde I, Escosa R, Aranda C, Huijben S, Mueller I, Marquès-Bonet T, Balloux F, Gilbert MTP, Lalueza-Fox C.
Molecular Biology and Evolution 2020;37(3);773-785
DOI: 10.1093/molbev/msz264
Detection of mutations associated with artemisinin resistance at k13- propeller gene and a near complete return of chloroquine susceptible falciparum malaria in Southeast of Tanzania
George M. Bwire, Billy Ngasala, Wigilya P. Mikomangwa, Manase Kilonzi & AppolinaryA. R. Kamuhabwa
Scientific Reports 2020;10;3500
DOI: 10.1038/s41598-020-60549-7
Assessment of Plasmodium falciparum drug resistance molecular markers from the Blue Nile State, Southeast Sudan
Mohamed AO, Hussien M, Mohamed A, Suliman A, Elkando NS, Abdelbagi H, Malik EM, Abdelraheem MH, Hamid MMA.
Malaria Journal 2020;19(1);78
DOI: 10.1186/s12936-020-03165-0
A molecular barcode to inform the geographical origin and transmission dynamics of Plasmodium vivax malaria
Diez Benavente E, Campos M, Phelan J, Nolder D, Dombrowski JG, Marinho CRF, Sriprawat K, Taylor AR, Watson J, Roper C, Nosten F, Sutherland CJ, Campino S, Clark TG.
PLoS Genetics 2020;16(2);e1008576
DOI: 10.1371/journal.pgen.1008576