The Ag1000G release of 765 Anopheles gambiae and coluzzii mosquito genomes is enabling investigation of these medically important malaria vector species at a resolution and breadth never possible before.
Perhaps the most striking genetic feature of these mosquitoes is the vast wealth of diversity found across their genomes – nearly one single nucleotide polymorphism (SNP) for every two bases of the reference genome. With this genetic diversity has come the discovery of many previously undetected non-synonymous (protein altering) mutations.
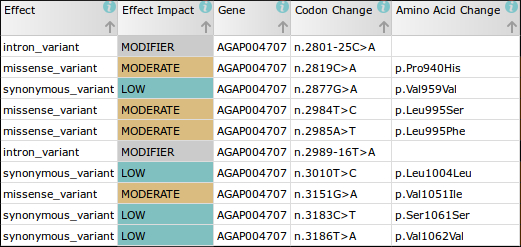
For example, in the first phase of the Ag1000G project we discovered 47 non-synonymous mutations in just the DNA sequence of the voltage gated sodium channel (Vgsc) gene alone. Most of these mutations have not been described before and, as the sequence encodes the protein target of pyrethroid and DDT insecticides, it is likely some play a role in insecticide resistance.
The trouble with borrowing nomenclature from other species
Some of the earliest discoveries of genetic mutations linked to insecticide resistance were not found in malaria vectors, but in other insects. Though mosquitoes may have orthologous genes containing equivalent resistance mutations to these insects, the actual amino acid sequences of the genes are likely to differ due to genetic divergence that occurred since their last common ancestor.
The nomenclature used to describe the positions of these mutations in the literature is therefore likely to be incorrect when applied to mosquitoes. For instance, the codon change in the Vgsc gene sequence which confers the kdr phenotype (resistance to pyrethroids and DDT), was discovered in the house fly (Musca domestica), where the associated codon change was 1014 (Vgsc-L1014F1). In An. gambiae, however, the equivalent insecticide associated codon position is actually at position 995 (Vgsc-L995F/S).
Other mutations, despite first being found in mosquitoes, were named relative to the orthologous genes from even more divergent organisms due to lack of available insect gene constructs at the time. The position of the Ace1-G119S mutation in the acetylcholinesterase gene, which confers resistance to carbamates and organophosphates, was discovered in mosquitoes2 where we now know its position is actually Ace1-G280S, but it was named according to the position in the Ace1 gene from a cartilaginous fish, the pacific electric ray (Torpedo californica).
With large numbers of mutations being discovered across many genes, using codon numbering from different species to describe the positions of new mutations becomes a hindrance. Also, as conservation of amino acid sequence is unlikely to be 100% between genes of even closely related species, there may be no equivalent position for some codons, and the nomenclature for these becomes unclear.
Using codon numbering from the An. gambiae gene-set
As more mosquitoes are sequenced and more genes implicated in insecticide resistance or other phenotypes are described, the number of potentially medically relevant mutations will continue to rise. More than ever before, future research into these vectors will require cataloguing these variants in a format that is both tractable and biologically relevant.
The Ag1000G Consortium has therefore decided to use the codon numbering from the An. gambiae gene-set for all genes and mutations, including those previously described in other species (such as Vgsc-L995F/S) and those newly-discovered.
This codon numbering is used in all Ag1000G outputs: data files included in public releases (e.g. variant effect annotations in VCF files), the Panoptes web application, and the papers that we're currently writing.
Download a codon numbering map
Insecticide resistance literature has historically used codon numbers from the housefly when describing Vgsc mutations; therefore, we’ve produced a codon numbering map between An. gambiae and Musca domestica for this gene. The map provides a quick reference of equivalent codons without the need to align gene sequences.
References
1. Williamson, M. S. et al. "Identification of mutations in the housefly para-type sodium channel gene associated with knockdown resistance (kdr) to pyrethroid insecticides." Molecular and General Genetics 252.1-2 (1996): 51-60.
2. Weill, M. et al. "Comparative genomics: Insecticide resistance in mosquito vectors." Nature 423.6936 (2003): 136-137.


