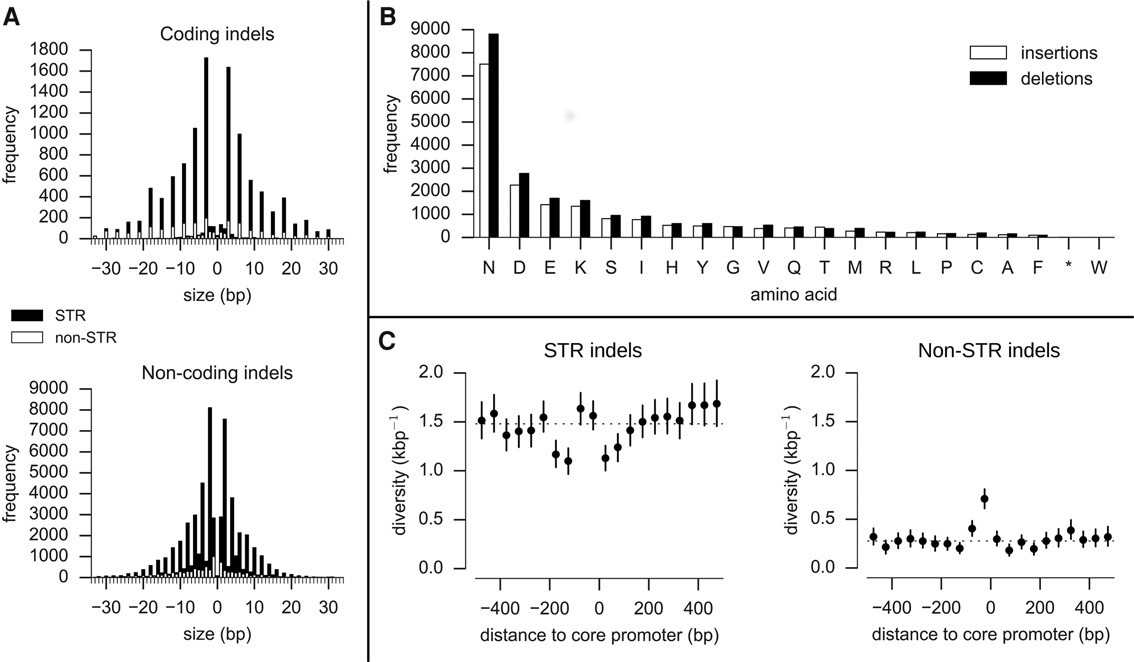
Figure 1. Properties of indels. (A) Indel size distribution (size > 0 are insertions, size < 0 are deletions). Solid black bars represent the frequency of indels that are expansions or contractions of short tandem repeats (STR); solid white bars represent the frequency of non-STR indels. Most coding indels are size multiples of 3, preserving the reading frame. Most noncoding indels are size multiples of 2, reflecting the abundance of poly(AT) repeats in noncoding regions. (B) Amino acids inserted and deleted (relative to the 3D7 reference genome). (C) Indel diversity in intergenic regions relative to the position of core promoters predicted by Brick et al. (2008). Each point represents the mean indel diversity in a 50-bp window at a given distance from the center of a core promoter. Vertical bars represent the 95% confidence interval from 1000 bootstraps. The dashed line is at the mean intergenic diversity for the given indel class (STR/non-STR).
By deep sequencing the 3D7 x HB3 (Walliker et al, 1987), HB3 x Dd2 (Wellems et al, 1990) and 7G8 x GB4 (Hayton et al, 2008) experimental crosses, the P. falciparum Genetic Crosses project has generated an open access resource for understanding natural genetic and phenotypic variation in P. falciparum.
A key finding is that indels are extremely abundant in the P. falciparum core genome:
“In most species, indels are relatively rare. For example, in the human genome there are roughly 10 SNPs for every indel,” says Alistair Miles from the Wellcome Trust Centre for Human Genetics, University of Oxford. “In P. falciparum there are more indels than SNPs, which is very unusual. Indels are therefore a major driver of genetic diversity and may be under-appreciated as a cause of phenotypic variation.”
The study also performed a high-resolution analysis of sexual recombination, confirming a high rate of crossover events and providing the first estimates for the rate of non-crossover events and the length of conversion tracts. Several recombination events were observed within copy number variants associated with drug resistance, providing evidence that sexual recombination could play an important role in the evolution of resistance.
“Professor Tom Wellems from the National Institute of Allergy and Infectious Diseases (NIAID), Maryland, USA, adds: “The analysis of the genomes of these three crosses provides us with unique insights into the biology of the parasite, giving us some indications on how the parasite evolves to face new challenges, such as the pressure from anti-malarial drugs.”
Learn more about the P. falciparum Crosses Project
Explore the P. falciparum crosses 1.0 data
Citation
Miles, Iqbal et al. Indels, structural variation, and recombination drive genomic diversity in Plasmodium falciparum. Genome Res. 2016 Aug 16. [Epub ahead of print].


