In the first continent-wide genomic study of malaria parasites in Africa, scientists have uncovered the genetic features of Plasmodium falciparum parasites that inhabit different regions of the continent, including the genetic factors that confer resistance to anti-malarial drugs. This sheds new light on the way that drug resistance is emerging in different locations and moving by various routes across Africa, putting previous success in controlling malaria at risk.
The research, published today (22 August) in Science, comes from the first network of African scientists, the Plasmodium Diversity Network Africa (PDNA), to work with genomic tools to study the diversity of malaria parasites across the continent. In collaboration with the Wellcome Sanger Institute, the researchers studied the genetic diversity of P. falciparum populations endemic to several countries in sub-Saharan Africa, including Ethiopia and Ghana. Genomic surveillance data will help to track the emergence and spread of drug-resistant strains, assisting efforts to eliminate malaria.
Malaria remains a global problem, with the deadliest parasite species P. falciparum prevalent across sub-Saharan Africa. Between 2000 and 2015, an ongoing drive to eliminate the disease has seen worldwide malaria deaths halve from 864,000 to 429,000 per year. In 2015, 92 per cent of global malaria deaths were in Africa, with 74 per cent of these occurring in children under five years of age.* But the findings of a new study suggest this progress may be at risk if new forms of treatment aren’t developed.
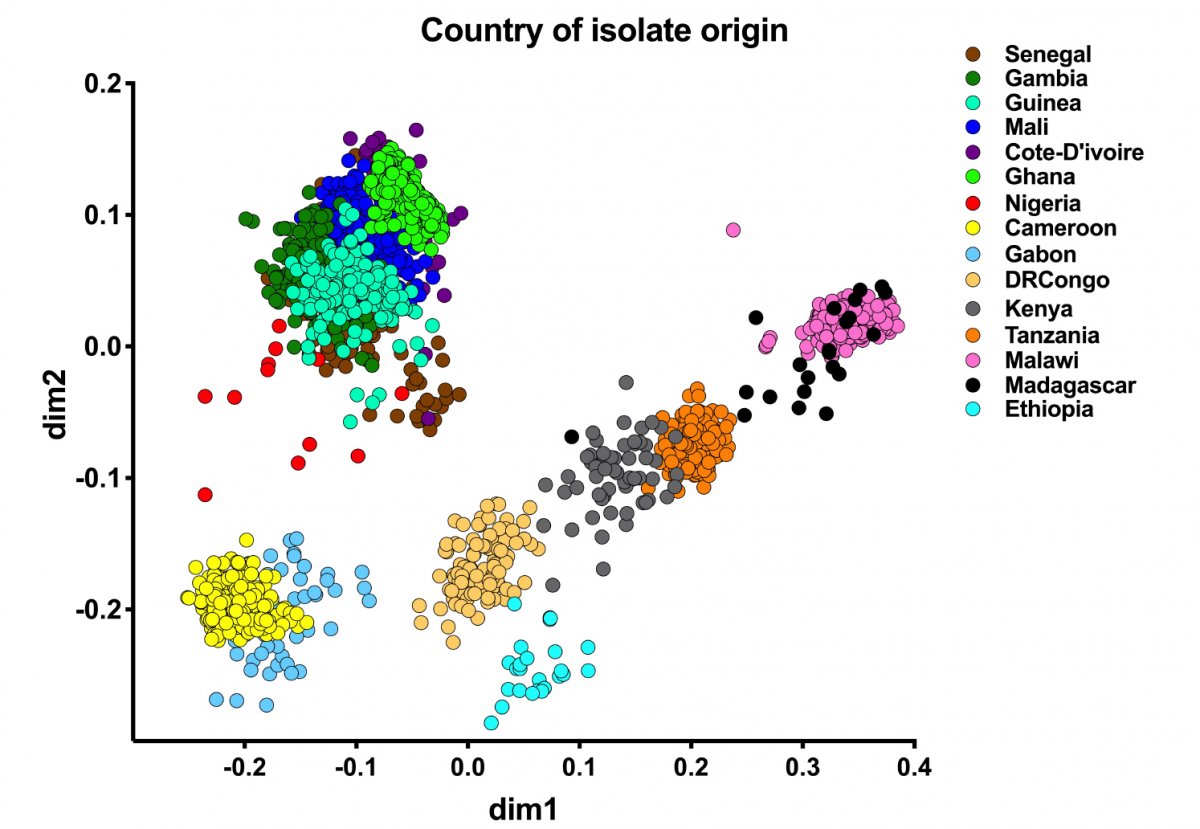
Although the population of P. falciparum parasites in sub-Saharan Africa is extremely genetically diverse, previous research suggested that this diversity was relatively similar across the continent. It was also thought that the flow of genetic material tended to be from east to west, with resistance to antimalarial drugs believed to originate in South East Asia.
The results of this new study indicate, however, that P. falciparum parasites are genetically distinct according to which region of Africa they are found. Furthermore, researchers found that these regional populations are sharing genetic material in all directions – including genes that can confer resistance to antimalarial drugs, with new types of drug resistance emerging in different parts of Africa. It is thought human migration, including that resulting from colonial activity, has played a part in the evolution of P. falciparum in Africa.
Samples of P. falciparum were collected from 15 African countries by PDNA and their genomes sequenced at the Wellcome Sanger Institute as part of the MalariaGEN data-sharing network**. The genetic data on these samples, along with other African data that have previously been generated and openly released by MalariaGEN, were analysed to trace ancestral connectivity between the various parasite populations.
Professor Abdoulaye Djimdé, Wellcome International Fellow at the Wellcome Sanger Institute and Chief of the Molecular Epidemiology and Drug Resistance Unit at the Malaria Research and Training Centre, University of Bamako said “Contrary to previous studies, we identified distinct Western, Central and Eastern populations of P. falciparum, as well as a highly-divergent Ethiopian population. Genetic material originating from all directions was shared by all populations, indicating that the flow of genes is multi-directional, as opposed to unidirectional from east to west as previously thought. This is crucial information for understanding how resistance to malaria drugs is developing in Africa.”
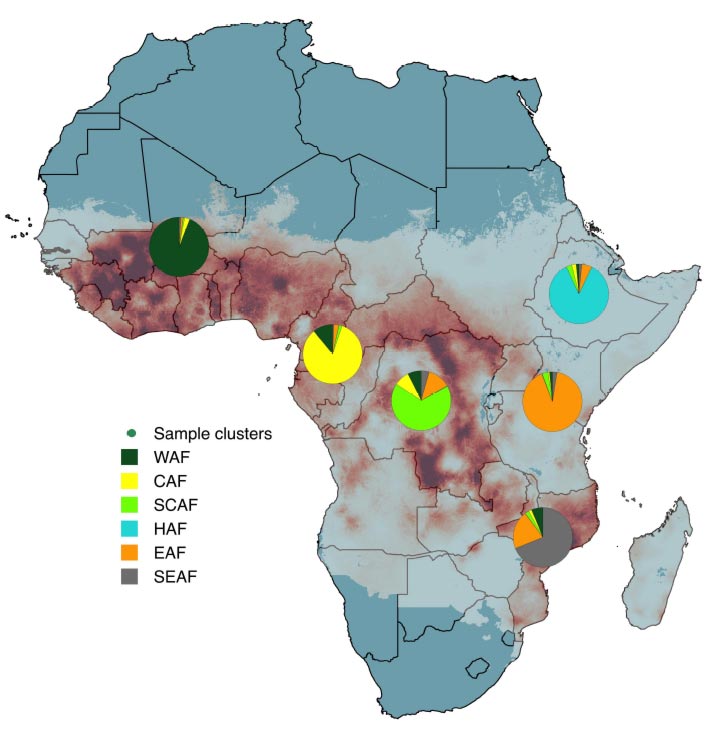
Researchers noted the fact that the Ethiopian parasite population is highly-differentiated from those in the rest of Africa, which suggested the ancestry of malaria parasites may have been influenced by human migration. The human population in Ethiopia also has a distinct ancestry to others in Africa, suggesting that the lack of colonization of the country might explain its outlier status. By contrast, parasites from distant former French colonies share genetic material.
The results confirmed that populations of P. falciparum have shared genetic information over time, particularly genes associated with resistance to antimalarial drugs.
Most concerningly, strong genetic signatures were detected on chromosome 12 in P. falciparum samples from Ghana and Malawi, raising the possibility that recent evolution of the parasite could compromise the effectiveness of artemisinin-based combination therapies (ACTs). ACTs combine multiple antimalarial drugs in one treatment to overcome resistance to one or more individual drugs.
Dr Alfred Amambua-Ngwa, first author of the study, a Wellcome International Fellow at the Wellcome Sanger Institute and Assistant Professor at the Medical Research Council Unit The Gambia, London School of Hygiene and Tropical Medicine said “Whatever the historic factors affecting the flow of genes between the distinct P. falciparum populations, the multi-directional flow we’ve identified raises the prospect of continental spread of resistance to artemisinin-based combination therapies, which could arise from anywhere in Africa. Genomic surveillance, and on a large scale, is going to be vital to track the emergence and spread of resistance to combination therapies.”
The establishment of the PDNA is an important step in continuing to track the spread of drug-resistant malaria in Africa at a crucial time, when efforts to eliminate the disease are now stalling and the prospect of multi-drug resistant strains of P. falciparum in Africa on the horizon.
Professor Dominic Kwiatkowski, Head of the Parasites and Microbes Programme at the Wellcome Sanger Institute, and Director of the MRC Centre for Genomics and Global Health says “Back in 2013, this impressive team of African scientists recognised the need to work together across the continent to monitor how this deadly malaria parasite is evolving. This has involved many practical and logistical challenges, and the team have done an amazing job in pulling together the most comprehensive genetic picture of the parasite population in different parts of Africa. It proves the feasibility of using modern genomic technologies to monitor malaria drug resistance in Africa, and it is now extremely important to ensure that the work is continued and used by policymakers and public health agencies to guide sustainable strategies for disease control.”
Michael Chew, Infection and Immunobiology Portfolio Manager at Wellcome says “This research could have implications for the future of malaria research and control in sub-Sahara Africa. By studying the genetic diversity of malaria across such a vast and diverse area, the research team have revealed the presence of key genetic differences within the malaria parasite P. falciparum strains across Africa.
“This could provide vital information into how drug-resistance is developing across the region and provides a stark reminder that the progress made in tackling malaria in sub-Sahara Africa is at risk of stalling unless we can develop new and effective treatments.”
Notes to Editors
*For more information on these figures and the state of malaria in general, see the WHO World Malaria Report 2016 https://www.who.int/malaria/media/world-malaria-report-2016/en/
**For more information on The Malaria Genomic Epidemiology Network (MalariaGEN), visit https://www.malariagen.net/about/
Link to paper:
https://science.sciencemag.org/cgi/doi/10.1126/science.aav5427
Funding:
This project was supported by the African Academy of Sciences (AAS), New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency), and Wellcome. For full funding information, please refer to the full study.
Press release courtesy of Wellcome Sanger Institute Press Office


