Plasmodium falciparum parasites are responsible for the majority of over 500,000 malarial deaths every year. An adaptive foe, these parasites can hide from the body’s immune system, cope with changes in the Anopheles (mosquito) vector, and develop resistance to antimalarial drugs, at a frightening rate.
Genomics is one of the most powerful tools available to observe these evolutionary processes in action in the parasites. Much of our early work studying natural genetic variation in Plasmodium parasites came about in collaboration with many different researchers around the world as part of the MalariaGEN P. falciparum Community Project. To date, this collaboration has built a catalogue of 1 million single nucleotide polymorphisms (SNPs) in more than 6,000 falciparum samples collected directly from malaria patients in Africa, Asia, Latin America and Oceania.
Using this rich data resource – the largest collection of Plasmodium genomes in the world – we are starting to understand the complex genetics of Plasmodium parasites. For example, the intricate genetic architecture underpinning resistance to the frontline drug, artemisinin.
But this is just the beginning! We are still far from a comprehensive and precise understanding of how this parasite evolves in in the wild and how we should respond to these constant changes. There are of course limitations with our current methods, but beyond that our view of genetic variation is primarily based on SNPs, leaving out other forms of variation such as indels. We are also not yet able to accurately detect changes in key regions of the Plasmodium genome including, for example, the hypervariable var genes, which contribute to the parasites’ ability to evade our immune system.
To generate a more complete, fine-grained view of genetic variation in Plasmodium parasites, we need solid reference genomes, good baseline data and reliable analytical methods. In short, we need to set the scene and lay solid foundations for future analyses.
These technical challenges are the key focus for the pilot phase of the Pf3k project, a global collaboration led by researchers at the Wellcome Trust Sanger Institute, the University of Oxford and the Broad Institute. Established within the past year, the Pf3k Consortium aims to analyse 3,000 P. falciparum samples from the major malaria-endemic regions of the world.
The overall aim is to provide a high-resolution view of natural variation in P. falciparum including those regions of the genome that are inaccessible using standard methods.
At the moment, we are very busy generating thousands of whole genomes from field samples that can act as high-quality reference genomes and assessing various methods to genotype them.
This is a big leap forward with respect to the current gold standard of using one reference, 3D7 v3, which is the whole genome sequence of a single parasite. This limits our ability to access the genome, particularly in regions that differ from the reference.
One good example is the challenges in genotyping crt, a clinically-significant gene involved in choloroquine resistance – and possibly with a role in emerging artemisinin resistance. This gene is so important that it remains one of the first places researchers tend to look.
The current reference has a very specific version of crt which is quite different from what we see in most genomes in Southeast Asia. And crt in Southeast Asia is again different from what we observe in other parts of the world. This geographical diversity makes aligning sequences from various parts of the world challenging; having reference genomes drawn from different populations will allow us to more readily compare like with like and, ultimately, increase the accuracy with which we can spot variants.
The Pf3k Consortium has prepared an initial data set comprising 2,375 samples sequenced here at the Sanger Institute as well as 137 samples from our colleagues at the Broad Institute in Boston, USA. This represents the full pilot set of samples, collected in major malaria-endemic regions in Africa and Asia.
Reflecting our commitment to the early and open release of data, earlier this month, the Pf3k Consortium made this large data set public, including sample information, accession numbers, analysis BAMs and preliminary genotypes. As with previous Pf3k data releases, these data are made available under Fort Lauderdale conditions and can be downloaded or explored using a user-friendly web application designed by colleagues at the MRC Centre for Genomics and Global Health.
Our attention is now focused on evaluating the methods used to generate this baseline data. Often optimised for human genomes, we need to understand to what degree these methods can be used straight off-the-shelf to analyse the Plasmodium genome, which differs in many ways from ours.
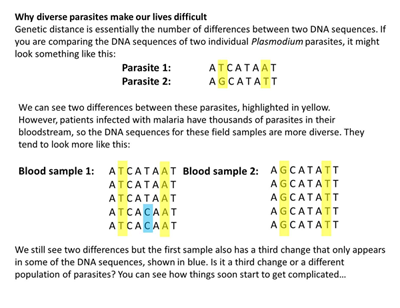
It may sound surprising but even some basic concepts, like allele frequencies and genetic distance, are not straightforward when dealing with Plasmodium genomes. When samples come directly from a patient, we’re not getting a single parasite – we get a population of parasites. Depending on a variety of ecological and epidemiological factors, these populations may be so inbreed as to actually appear as a single genome (clonal sample) or may be very diverse (mixed infections).
A funny consequence of mixed infection is that some Plasmodium genomes look like they have an extra set of chromosomes at certain positions! To further increase the complexity, in areas where other Plasmodium parasites are co-endemic, these populations might even be made of different species.
As we improve the resolution and accuracy of our analyses of genetic variation, we’ll be able to delve deeper into key scientific questions like how populations of Plasmodium parasites are evolving, migrating to different locations and developing drug resistance.
Roberto Amato is a Research Associate in Statistical Genomics who is involved in the analysis of natural genetic variation in the Plasmodium parasites that cause malaria. His primary focus is on developing new statistical methods to understand the evolution of these parasites at a population level, in order to shed light on the underlying genetics of antimalarial drug resistance. Based in the Wellcome Trust Sanger Institute’s Malaria Programme, Roberto works closely with colleagues at the Wellcome Trust Centre for Human Genetics at University of Oxford and supports several global collaborations including the MalariaGEN P. falciparum Community Project and Pf3k.
References
- Miotto O, Amato R, et al. (2015). Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nature Genetics. DOI:10.1038/ng.3189
- Miotto O, et al (2013). Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nature Genetics. DOI:10.1038/ng.2624
- Claessens A, Hamilton WL, et al (2014). Generation of antigenic diversity in Plasmodium falciparum by structured rearrangement of Var genes during mitosis. PLOS Genetics DOI:10.1371/journal.pgen.1004812

