Malaria parasites are constantly evolving to survive drug treatments deployed against them. To combat malaria effectively, monitoring the genetic changes they carry and sustain across different populations is essential.
MalariaGEN builds and shares the world’s largest open data resource of whole genome sequences for Plasmodium parasites. However, its size and complexity can present a challenge for researchers and public health officials who do not have the extensive bioinformatics expertise required to draw actionable insights from the data.
The Pf-HaploAtlas app is an accessible and intuitive platform for exploring and interpreting malaria genomic data. By providing detailed visualisations pulled from a comprehensive data resource, Pf-HaploAtlas empowers users to track genetic mutations in malaria parasites more effectively.
How does Pf-HaploAtlas work?
Pf-HaploAtlas enables any user with an internet connection to study and track genetic mutations across any gene in the P. falciparum core genome. It draws from the MalariaGEN Pf7 resource, a vast dataset of 16,203 quality-controlled samples collected from 33 countries between 1984 and 2018.
What are haplotypes?
Haplotypes are groups of DNA mutations that are inherited together. These variations are often located close to each other in the genetic code, making them more likely to be passed down together. Certain haplotypes include mutations that lead to drug resistance in malaria parasites and in some cases, it’s a specific combination of mutations that confers resistance. Studying haplotypes instead of individual single-nucleotide mutations allows us to track these more complicated mutation combinations. They also allow scientists to trace genetic linkages and inheritance patterns.
Learn to use Pf-HaploAtlas
Here's everything we can learn about kelch13 and aat1 using the Pf-HaploAtlas app
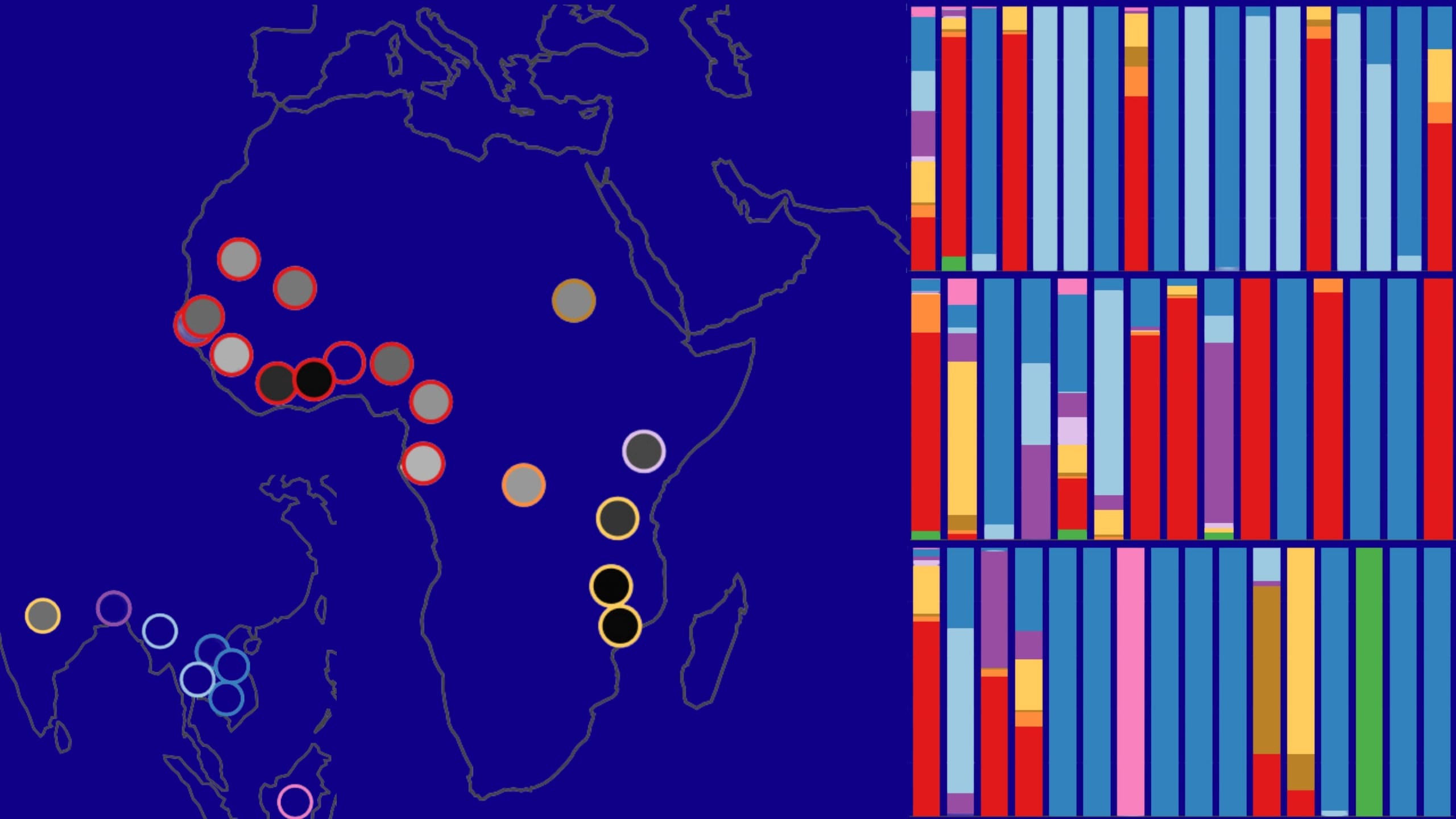
The app provides three innovative plots to visualise haplotypes for all core genes of the P. falciparum parasite: the UpSet, abacus, and world map plots. For each chosen haplotype, users can see the specific mutations present, explore their frequencies and distributions worldwide, and compare prevalence over time.
Developed using open data and open-source software, Pf-HaploAtlas is accessible to a wide range of users interested in malaria genomic surveillance and data analysis, including researchers and public health officials.
Apart from tracking known mutations linked to drug resistance, the app’s unique features can help identify emerging patterns in less-studied genes.
Amino acid haplotypes
The Pf-HaploAtlas focuses on haplotypes where the mutations change the amino acid being produced. This takes out some of the noise in the data created by single point mutations that result in no change to the amino acids and therefore proteins coded for. In general, protein-level analysis is more relevant for studying functional impacts like a parasite’s tolerance for antimalarial drugs.
Two genes for you to start exploring Pf-HaploAtlas
kelch13
kelch13 is a key gene involved in malaria parasites resisting artemisinin-based drugs, the WHO-recommended first-line treatment for malaria. While variants of P. falciparum parasites with kelch13 mutations were first discovered to be widespread in Southeast Asia, they have since been identified in several African countries. Look at the rise of clinically important kelch13 haplotypes for yourself using the app.
aat1
The aat1 gene encodes an amino acid transporter protein in the digestive vacuole membrane of P. falciparum. Aat1 mutations have recently been linked to parasites resisting the anti-malarial drug chloroquine. Researchers who analysed the MalariaGEN Pf6 dataset also found different aat1 mutations from Africa and Asia. Try searching for potential patterns of selection in this less-studied gene.
A step closer to decentralised malaria genomic surveillance
Pf-HaploAtlas is designed to grow with advancements in malaria genomics. Future updates will incorporate the latest data from MalariaGEN data releases and the app will continue to be a relevant tool to track and discover recent genetic changes.
The app can also integrate new functionalities based on user feedback. With the potential to expand to other malaria parasites like Plasmodium vivax, continuous improvement will help ensure that Pf-HaploAtlas remains a vital tool for genomic surveillance and malaria control efforts.
Pf-HaploAtlas is a simplified, user-friendly platform for anyone interested in malaria genomics to visualise mutations in target genes and how they’ve changed in populations over time. Professor Olivo Miotto, University of Oxford and MORU, Bangkok.
As more malaria researchers and intervention programs delve into the P. falciparum whole genome data using Pf-HaploAtlas, it could spur discoveries of lesser-well-studied genetic mutations that play a role in drug resistance and evasion of other interventions. Professor Alfred Amambua-Ngwa, MRC Unit The Gambia at LSHTM.
Useful links
- App: https://apps.malariagen.net/pf-haploatlas
- Manuscript DOI: https://doi.org/10.1093/bioinformatics/btae673
- Contact us to report a bug, request a feature, or provide feedback: https://forms.gle/mDwYr2cPL37dDzPs6
- When publishing work that uses data and/or plots from Pf-HaploAtlas, please cite the following:
Lee C, Ünlü ES, White NFD, Almagro-Garcia J, Ariani C, Pearson RD. Pf-HaploAtlas: An interactive web app for spatiotemporal analysis of P. falciparum genes. BioRxiv 603783 [Preprint] July 16, 2024. Available from: https://doi.org/10.1101/2024.07.16.603783