Kicking things off - The Malaria Vector Genome Observatory
Community-driven development is key to sustainable progress.
This is the message Anastasia Hernandez-Koutoucheva (Wellcome Sanger Institute, UK) had for delegates as she began the first session. She presented the Malaria Vector Genome Observatory (Vobs), an open-to-all source of vector genomic data inspired by the Ag1000G project.
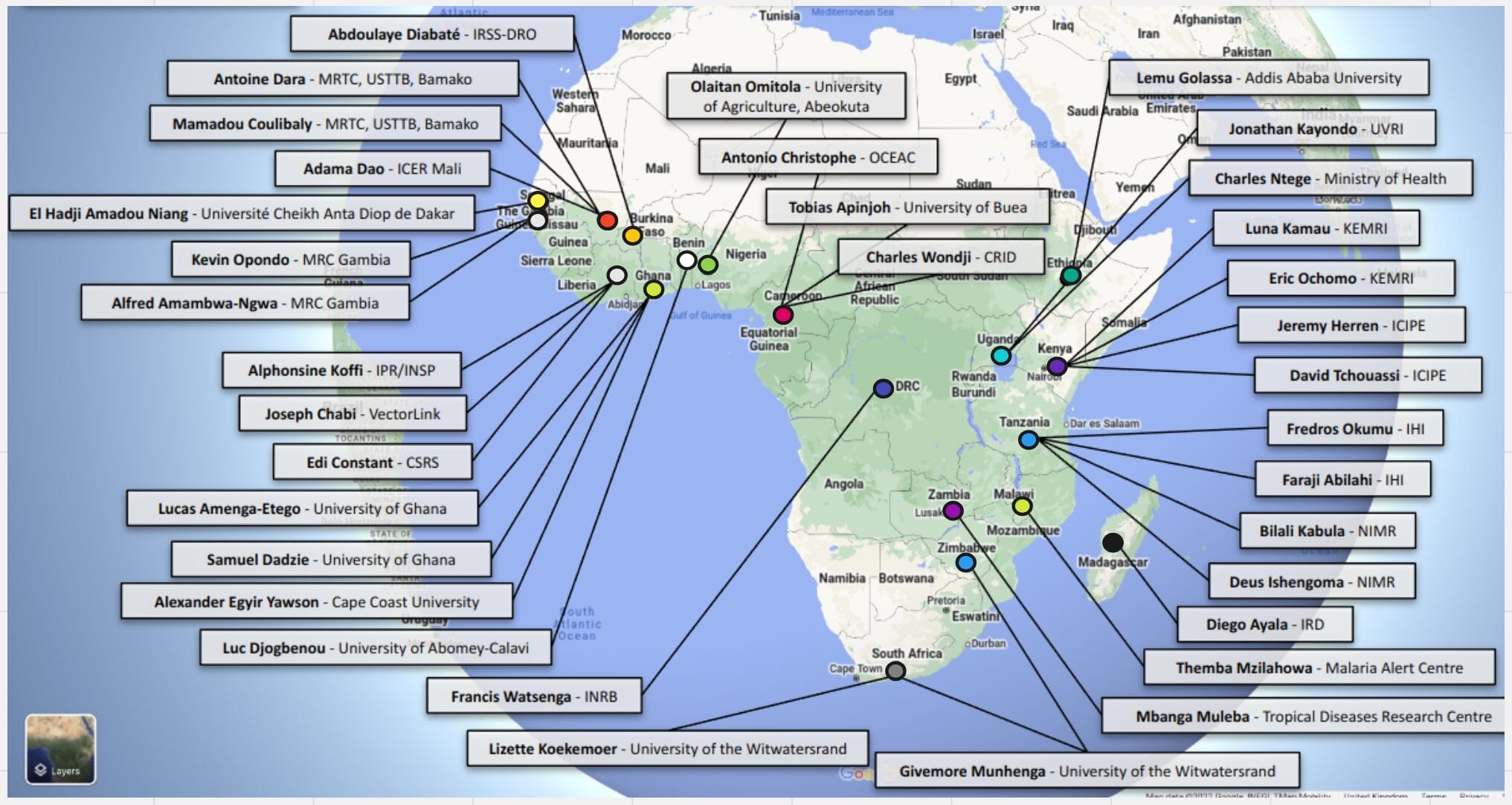
She shared an impressive list of partners from around Africa who have contributed to the project. “In 2018 a brief was sent around asking for people to contribute every mosquito that bites them,” she said. “Behind each of these labels, there is a whole team of people who have sent their mosquitos and data to the observatory.”
The Vector Observatory now contains over 25,000 mosquito samples and as it continues to grow, Anastasia detailed the commitment to open data sharing. She also announced the brand new vobs updates webpage which provides regular, transparent information about any changes to and results from the observatory.
The impacts of the Vector Observatory can already be seen in many of the presentations given at GEM, where the data was referenced for many research projects. For example, drawing influence from the MalariaGEN Ag1000G project, Jacob Tennesen (Harvard School of Public Health) shared insights from the Ad1000G project, providing a comprehensive evolutionary analysis of Anopheles darlingi in South America.
As the Observatory grows, we can also look forward to the addition of new data.
Early careers spotlight
This edition of the GEM Conference brought together many new collaborators in the malaria genomics community.
Being the first GEM conference to be held in person since 2018, the audience was filled with many first-time attendees who brought new insights and passion, showing the great opportunity the conference provides for early career researchers to enrich collaboration. Read about two insightful cases below.
What changes are needed in low transmission settings?

Uganda is a country that covers over 241,000 km2 and, like many malaria-endemic countries, transmission varies across geographical areas. This means that a single control strategy cannot be used to effectively control malaria in the country as a whole and instead, we must adapt our efforts to match the specific needs of each area.
This is the thought-provoking message Monica Mbabazi (Makerere University, Uganda) presented. She had performed a characterisation of local cases and factors sustaining malaria transmission in Southwestern Uganda, a region known for very low transmission. Research like this helps inform the National Malaria Control Division’s work towards the goal of regional elimination. By investigating travel history and malaria genomics, Monica showed the different driving factors behind transmission and that different sites require different interventions to control malaria.
Ancient DNA – What can history tell us?
The second day began with Megan Michel (Max Planck, Germany) exploring the history of human malaria using ancient DNA. A look at the evolutionary history of common Plasmodium species revealed that P. falciparum and P. vivax’s closest relatives are parasites affecting great apes. Similarly, by examining the prevalence of human alleles selected for by pathogens, Michel explained that “we can use these markers as biomolecular footprints to trace the presence of these parasites in the past”.
A great example of this is the prevalence of the sickle cell allele and P. falciparum incidence across sub-Saharan Africa. She showed how ancient DNA can be recovered from historical remains and how a collection of ancient DNA samples can be used to examine historical malaria cases.
Getting Interactive - Fighting arteminisin resistance in ‘GEMia’
What is it like to be a public health professional and make the decisions of where to send our treatment resources?

This was the question Kathryn Murie and Dr Amy Wesolowki wanted the GEM delegates to consider as they transported the crowd into the shoes of a public health official. With the growing pressures of artemisinin resistance in Africa, delegates examined the aggregate site data, built an analysis workflow, and output frequency estimations. This work informed the final response of where to send monoclonal antibodies and resulted in the effective administration of treatment to the regions most in need!
Delegates then heard from Dr Shazia Ruybal-Pesántez (Imperial College London) and Dr Francis Chikuse (Africa CDC), who highlighted the importance of open genomic data sharing and shared a proposed protocol for harmonisation from the Africa Centre for Disease Control. This session proved a great opportunity to recognise the impact of data analysis and sharing for public health.
What does it take to eliminate malaria?
Dr Dionica Gamboa (Universidad Peruana Cayetano Heredia, Peru), provided a detailed overview of molecular surveillance efforts in Peru. Often surveillance efforts in Peru focus on the higher case incidence within Loreto but equally important is elimination in hard-to-reach communities. Her work has included a study involving an indigenous community where prevalence is high. Studies such as this will no doubt be essential to ensure complete elimination.
Another country making significant progress incorporating genomic information into public health decision-making is Vietnam, where they experience about 500 malaria cases annually. Dr Nguyen Thanh Thuy Nhien (Oxford University Clinical Research Unit, Vietnam) shared the challenges the country now faces to reach their goal of elimination by 2030. These include increases in drug resistant parasites, insecticide resistant vectors, and changes in the proportions of non-falciparum malaria. Sentinel sites have been used across the country to collect surveillance data from clinical trials allowing for sequencing of 30-40% of annual samples. With this data, Dr Thuy Nhien has identified notable findings such as decreases in P. vivax diversity in recent years and local variation in specific gene mutations associated with drug resistance. Her team’s work continues to be available to inform ongoing national malaria control efforts.
Lastly, Dr Hoseah Akala (Kenya Medical Research Institute/Walter Reed Army Institute of Research - Africa, Kenya) provided an update on the burden of non-falciparum species and drug resistance profiles of Plasmodium Ovale. P. ovale provides an example of an increasing species burden as P. falciparum incidence has fallen due to drug policies over time. In addition to this, Hoseah and colleagues have identified a high prevalence of mutations in the dhfr gene in mosquitoes. This finding was linked to the selective pressure created by sulfadoxine-pyrimethamine use. He highlighted the importance of researching and addressing these non-falciparum species to further control malaria.
Advancing our goal
To end the conference, audience members got involved in a panel discussion chaired by Abdoulaye Djimdé (University of Bamako, Mali). Members discussed the steps needed to achieve malaria elimination, alongside topics such as the use of data to inform treatment policy and the implementation of whole genome sequencing (WGS) data into routine clinical practice.
GEM 2024 also hosted the inaugural presentation of the ‘Dominic Kwiatkowski GEM award’, awarded for outstanding contributions to the malaria genomics field. The awardees, Dr Eniyou Cheryll Oriero and Dr Moses Ikegbunam, have shared their vision and hopes for the future of the field.
The conference displayed the outstanding research being undertaken within the malaria genomics field and highlighted the bright future in achieving our collective goal of malaria elimination.
Online attendees can access recordings and poster materials online until Wednesday 16th October.



